You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001416_00754
You are here: Home > Sequence: MGYG000001416_00754
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cellulomonas massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas massiliensis | |||||||||||
| CAZyme ID | MGYG000001416_00754 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 814020; End: 816095 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH13 | 71 | 335 | 1.1e-101 | 0.9962825278810409 |
| CBM20 | 599 | 680 | 1.8e-31 | 0.9111111111111111 |
| CBM25 | 491 | 564 | 7.5e-25 | 0.9615384615384616 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd11317 | AmyAc_bac_euk_AmyA | 3.32e-157 | 46 | 391 | 1 | 329 | Alpha amylase catalytic domain found in bacterial and eukaryotic Alpha amylases (also called 1,4-alpha-D-glucan-4-glucanohydrolase). AmyA (EC 3.2.1.1) catalyzes the hydrolysis of alpha-(1,4) glycosidic linkages of glycogen, starch, related polysaccharides, and some oligosaccharides. This group includes AmyA proteins from bacteria, fungi, mammals, insects, mollusks, and nematodes. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| cd11315 | AmyAc_bac1_AmyA | 6.97e-58 | 46 | 393 | 1 | 347 | Alpha amylase catalytic domain found in bacterial Alpha-amylases (also called 1,4-alpha-D-glucan-4-glucanohydrolase). AmyA (EC 3.2.1.1) catalyzes the hydrolysis of alpha-(1,4) glycosidic linkages of glycogen, starch, related polysaccharides, and some oligosaccharides. This group includes Firmicutes, Proteobacteria, Actinobacteria, and Cyanobacteria. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| cd05808 | CBM20_alpha_amylase | 5.40e-42 | 599 | 691 | 2 | 95 | Alpha-amylase, C-terminal CBM20 (carbohydrate-binding module, family 20) domain. This domain is found in several bacterial and fungal alpha-amylases including the maltopentaose-forming amylases (G5-amylases). Most alpha-amylases have, in addition to the C-terminal CBM20 domain, an N-terminal catalytic domain belonging to glycosyl hydrolase family 13, which hydrolyzes internal alpha-1,4-glucosidic bonds in starch and related saccharides, yielding maltotriose and maltose. Two types of soluble substrates are used by alpha-amylases including long substrates (e.g. amylose) and short substrates (e.g. maltodextrins or maltooligosaccharides). The CBM20 domain is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
| pfam00686 | CBM_20 | 5.85e-35 | 599 | 688 | 2 | 95 | Starch binding domain. |
| cd05467 | CBM20 | 2.51e-33 | 600 | 690 | 2 | 96 | The family 20 carbohydrate-binding module (CBM20), also known as the starch-binding domain, is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARK04721.1 | 0.0 | 45 | 691 | 73 | 727 |
| QDP76525.1 | 0.0 | 45 | 691 | 73 | 727 |
| QUB99931.1 | 0.0 | 45 | 691 | 73 | 726 |
| QDW64271.1 | 0.0 | 36 | 691 | 47 | 701 |
| QJW35995.1 | 0.0 | 45 | 691 | 82 | 734 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1G94_A | 1.46e-113 | 50 | 472 | 6 | 442 | CRYSTALSTRUCTURE ANALYSIS OF THE TERNARY COMPLEX BETWEEN PSYCHROPHILIC ALPHA AMYLASE FROM PSEUDOALTEROMONAS HALOPLANCTIS IN COMPLEX WITH A HEPTA-SACCHARIDE AND A TRIS MOLECULE [Pseudoalteromonas haloplanktis],1G9H_A TERNARY COMPLEX BETWEEN PSYCHROPHILIC ALPHA-AMYLASE, COMII (PSEUDO TRI-SACCHARIDE FROM BAYER) AND TRIS (2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL) [Pseudoalteromonas haloplanktis],1L0P_A Crystal Structure Analysis Of The Complex Between Psychrophilic Alpha Amylase From Pseudoalteromonas Haloplanctis And Nitrate [Pseudoalteromonas haloplanktis] |
| 1JD9_A | 1.71e-113 | 50 | 472 | 6 | 442 | ChainA, ALPHA-AMYLASE [Pseudoalteromonas haloplanktis] |
| 1AQH_A | 1.71e-113 | 50 | 472 | 6 | 442 | ALPHA-AMYLASEFROM ALTEROMONAS HALOPLANCTIS [Pseudoalteromonas haloplanktis],1AQM_A ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS COMPLEXED WITH TRIS [Pseudoalteromonas haloplanktis],1B0I_A ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS [Pseudoalteromonas haloplanktis] |
| 1JD7_A | 2.41e-113 | 50 | 472 | 6 | 442 | ChainA, ALPHA-AMYLASE [Pseudoalteromonas haloplanktis] |
| 1KXH_A | 8.10e-113 | 50 | 472 | 6 | 442 | ChainA, alpha-amylase [Pseudoalteromonas haloplanktis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22998 | 2.35e-176 | 31 | 691 | 17 | 568 | Alpha-amylase OS=Streptomyces violaceus OX=1936 GN=aml PE=2 SV=1 |
| P09794 | 9.04e-170 | 32 | 691 | 21 | 565 | Alpha-amylase OS=Streptomyces limosus OX=1947 GN=aml PE=3 SV=1 |
| P30270 | 1.02e-168 | 32 | 691 | 21 | 565 | Alpha-amylase OS=Streptomyces griseus OX=1911 GN=amy PE=3 SV=1 |
| P27350 | 1.55e-147 | 31 | 477 | 13 | 459 | Alpha-amylase OS=Streptomyces thermoviolaceus OX=1952 GN=amy PE=3 SV=2 |
| P29750 | 5.88e-130 | 32 | 691 | 25 | 605 | Alpha-amylase OS=Thermomonospora curvata OX=2020 GN=tam PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
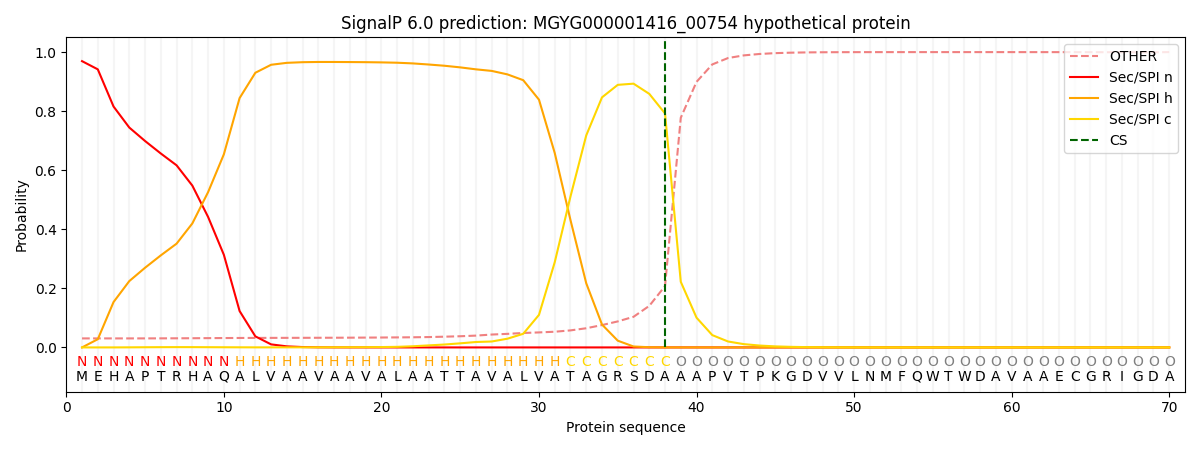
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.034005 | 0.964450 | 0.000344 | 0.000573 | 0.000320 | 0.000296 |