You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_02623
You are here: Home > Sequence: MGYG000001422_02623
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_02623 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1318965; End: 1320311 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 315 | 442 | 1e-16 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08522 | DUF1735 | 4.48e-14 | 182 | 286 | 15 | 120 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam00754 | F5_F8_type_C | 5.14e-14 | 320 | 442 | 5 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam08522 | DUF1735 | 7.71e-10 | 33 | 157 | 1 | 120 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam07738 | Sad1_UNC | 5.19e-04 | 387 | 446 | 62 | 128 | Sad1 / UNC-like C-terminal. The C. elegans UNC-84 protein is a nuclear envelope protein that is involved in nuclear anchoring and migration during development. The S. pombe Sad1 protein localizes at the spindle pole body. UNC-84 and and Sad1 share a common C-terminal region, that is often termed the SUN (Sad1 and UNC) domain. In mammals, the SUN domain is present in two proteins, Sun1 and Sun2. The SUN domain of Sun2 has been demonstrated to be in the periplasm. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT88787.1 | 2.14e-239 | 1 | 448 | 1 | 447 |
| BCA52367.1 | 5.42e-185 | 1 | 447 | 1 | 446 |
| QUT73146.1 | 5.42e-185 | 1 | 447 | 1 | 446 |
| QUT40999.1 | 4.42e-184 | 1 | 447 | 1 | 446 |
| QGT72760.1 | 1.36e-171 | 7 | 448 | 10 | 449 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GGL_A | 2.66e-25 | 302 | 430 | 12 | 140 | X-RayStructure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. [Bacteroides thetaiotaomicron],6OE2_A X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. Re-refinement of 3GGL with correct metal Mn replacing Zn. New metal confirmed with PIXE analysis of original sample. [Bacteroides thetaiotaomicron] |
| 2KD7_A | 2.81e-25 | 301 | 430 | 1 | 130 | ChainA, Putative chitobiase [Bacteroides thetaiotaomicron VPI-5482] |
| 3F2Z_A | 1.71e-22 | 302 | 435 | 2 | 135 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
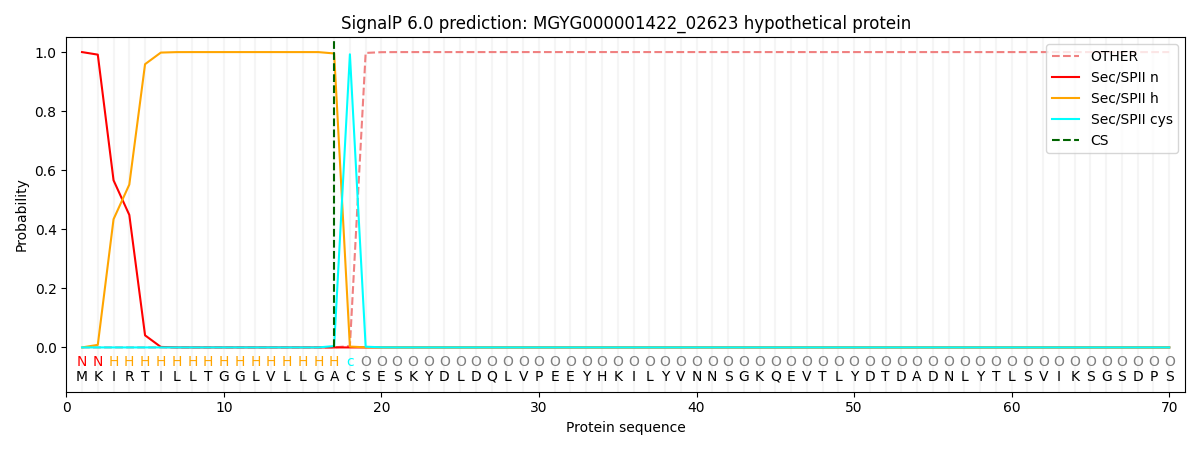
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000062 | 0.000000 | 0.000000 | 0.000000 |
