You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_03686
Basic Information
help
| Species |
Bacteroides oleiciplenus
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus
|
| CAZyme ID |
MGYG000001422_03686
|
| CAZy Family |
GH10 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001422 |
7071122 |
Isolate |
not provided |
Asia |
|
| Gene Location |
Start: 381088;
End: 383301
Strand: +
|
| Family |
Start |
End |
Evalue |
family coverage |
| GH10 |
503 |
733 |
7.5e-18 |
0.6831683168316832 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG3693
|
XynA |
3.68e-04 |
480 |
734 |
105 |
339 |
Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
This protein is predicted as LIPO
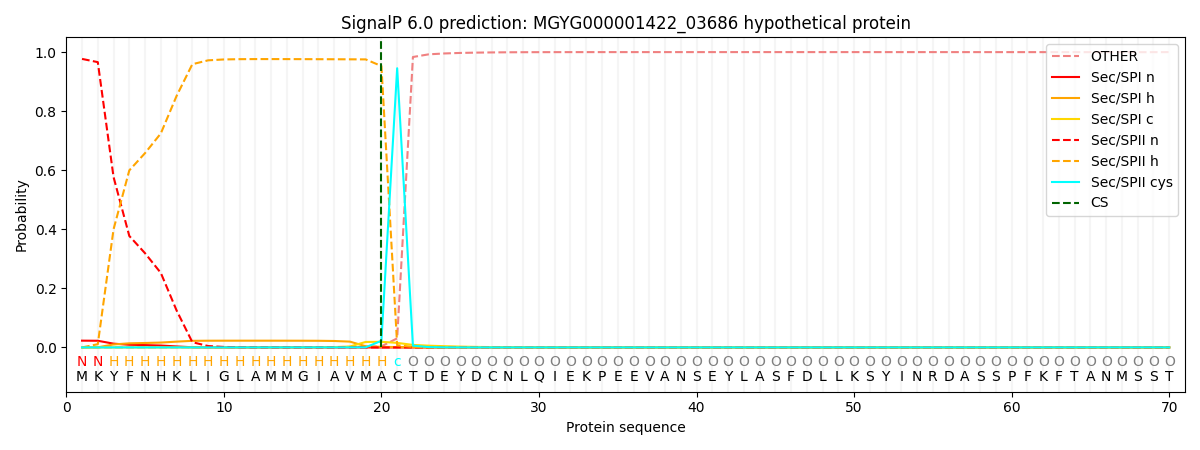
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000315
|
0.021875
|
0.977747
|
0.000019
|
0.000031
|
0.000024
|
There is no transmembrane helices in MGYG000001422_03686.

