You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001429_01072
You are here: Home > Sequence: MGYG000001429_01072
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A ihumii | |||||||||||
| CAZyme ID | MGYG000001429_01072 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11894; End: 13444 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 23 | 370 | 1.3e-49 | 0.8586118251928021 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03901 | Glyco_transf_22 | 6.25e-23 | 17 | 342 | 1 | 337 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| PLN02816 | PLN02816 | 1.72e-13 | 8 | 335 | 31 | 353 | mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL06999.1 | 5.90e-127 | 15 | 505 | 15 | 506 |
| AOW16884.1 | 1.01e-55 | 17 | 342 | 11 | 331 |
| AUR51123.1 | 1.57e-53 | 16 | 337 | 10 | 326 |
| ANI89374.1 | 1.91e-51 | 23 | 350 | 21 | 344 |
| QVY67119.1 | 1.15e-50 | 27 | 337 | 21 | 326 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q94A15 | 1.60e-14 | 8 | 329 | 31 | 347 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
| Q9JJQ0 | 1.99e-08 | 17 | 335 | 52 | 362 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
| Q753C1 | 4.04e-07 | 23 | 286 | 12 | 283 | GPI mannosyltransferase 4 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=SMP3 PE=3 SV=3 |
| Q1LZA0 | 5.65e-07 | 38 | 343 | 74 | 371 | GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1 |
| Q92521 | 9.24e-06 | 38 | 335 | 84 | 373 | GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000058 | 0.000005 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
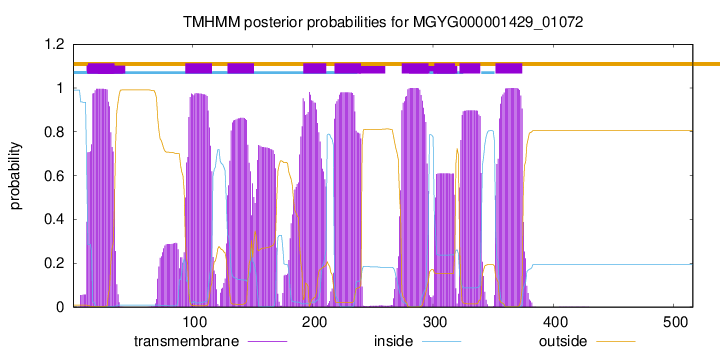
| start | end |
|---|---|
| 13 | 35 |
| 94 | 116 |
| 129 | 151 |
| 192 | 211 |
| 218 | 240 |
| 274 | 296 |
| 301 | 318 |
| 322 | 339 |
| 352 | 374 |
