You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001433_02733
You are here: Home > Sequence: MGYG000001433_02733
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides salyersiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides salyersiae | |||||||||||
| CAZyme ID | MGYG000001433_02733 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 460711; End: 463419 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 68 | 255 | 2.1e-78 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17161 | DUF5123 | 5.50e-12 | 683 | 819 | 1 | 116 | Domain of unknown function (DUF5123). |
| pfam16318 | DUF4957 | 8.20e-11 | 531 | 664 | 1 | 141 | Domain of unknown function (DUF4957). This family consists of uncharacterized proteins around 150 residues in length and is mainly found in various Bacteroides and Prevotella species. The function of this protein is unknown. |
| smart00656 | Amb_all | 2.40e-06 | 62 | 255 | 3 | 186 | Amb_all domain. |
| TIGR04183 | Por_Secre_tail | 2.47e-05 | 843 | 901 | 11 | 71 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| pfam18962 | Por_Secre_tail | 7.08e-04 | 843 | 901 | 9 | 72 | Secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber have on average twenty or more copies of this C-terminal domain, associated with sorting to the outer membrane and covalent modification. This domain targets proteins to type IX secretion systems and is secreted then cleaved off by a C-terminal signal peptidease. Based on similarity to other families it is likely that this domain adopts an immunoglobulin like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT75603.1 | 0.0 | 1 | 902 | 1 | 902 |
| QUT77823.1 | 4.11e-265 | 15 | 494 | 51 | 524 |
| QUT93111.1 | 9.81e-260 | 8 | 485 | 37 | 511 |
| ALJ61309.1 | 9.20e-259 | 8 | 485 | 41 | 515 |
| QDO69223.1 | 6.84e-255 | 15 | 485 | 43 | 511 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 3.75e-47 | 2 | 485 | 7 | 414 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q2UB83 | 2.06e-45 | 2 | 485 | 7 | 414 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| A1DPF0 | 6.13e-44 | 9 | 485 | 15 | 415 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
| Q4WL88 | 1.54e-43 | 9 | 485 | 15 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| B0XMA2 | 2.83e-43 | 9 | 485 | 15 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
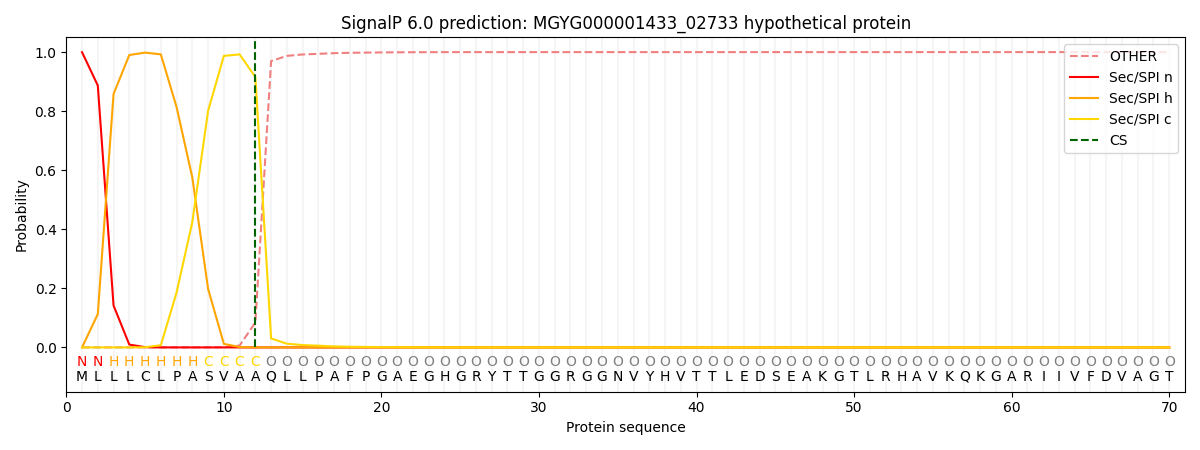
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000888 | 0.998254 | 0.000202 | 0.000238 | 0.000195 | 0.000197 |
