You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001488_01014
You are here: Home > Sequence: MGYG000001488_01014
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
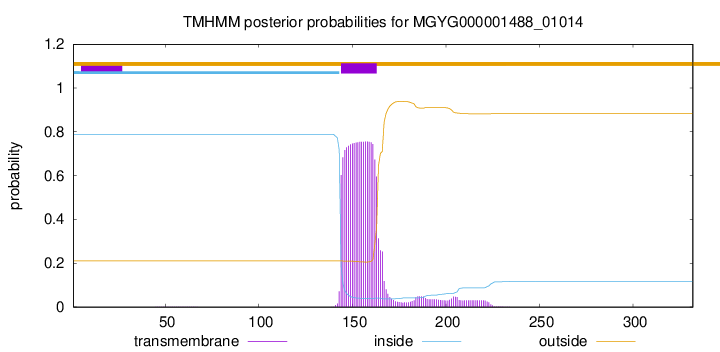
TMHMM annotations
Basic Information help
| Species | Risungbinella massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Thermoactinomycetales; Thermoactinomycetaceae; Risungbinella; Risungbinella massiliensis | |||||||||||
| CAZyme ID | MGYG000001488_01014 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 900394; End: 901392 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 42 | 225 | 1.4e-31 | 0.7444933920704846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3509 | LpqC | 5.44e-60 | 43 | 330 | 46 | 309 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam10503 | Esterase_phd | 1.52e-58 | 46 | 245 | 3 | 199 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| TIGR01840 | esterase_phb | 1.60e-40 | 46 | 243 | 1 | 196 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| COG1506 | DAP2 | 1.66e-14 | 40 | 229 | 372 | 565 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG4099 | COG4099 | 6.18e-13 | 46 | 327 | 176 | 386 | Predicted peptidase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCG71769.1 | 6.27e-43 | 61 | 325 | 65 | 307 |
| AXI86997.1 | 1.21e-41 | 49 | 298 | 42 | 277 |
| QBQ61857.1 | 1.28e-40 | 49 | 298 | 57 | 291 |
| QGL48637.1 | 2.29e-40 | 49 | 298 | 55 | 287 |
| QOV46954.1 | 3.37e-40 | 49 | 298 | 54 | 289 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5X6S_A | 1.38e-16 | 48 | 242 | 21 | 218 | Acetylxylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
| 3WYD_A | 2.03e-12 | 46 | 243 | 23 | 181 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 3DOH_A | 4.31e-07 | 46 | 175 | 160 | 297 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P52090 | 2.24e-38 | 49 | 316 | 56 | 301 | Poly(3-hydroxyalkanoate) depolymerase C OS=Paucimonas lemoignei OX=29443 GN=phaZ1 PE=3 SV=1 |
| Q5B037 | 1.01e-21 | 46 | 298 | 47 | 287 | Acetylxylan esterase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=axeA PE=1 SV=2 |
| A1DBP9 | 3.26e-21 | 15 | 242 | 24 | 248 | Probable acetylxylan esterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=axeA PE=3 SV=1 |
| Q8NJP6 | 4.58e-20 | 43 | 242 | 49 | 251 | Acetylxylan esterase A OS=Talaromyces purpureogenus OX=1266744 GN=axeA PE=1 SV=1 |
| Q4WBW4 | 1.44e-19 | 15 | 242 | 24 | 248 | Probable acetylxylan esterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=axeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
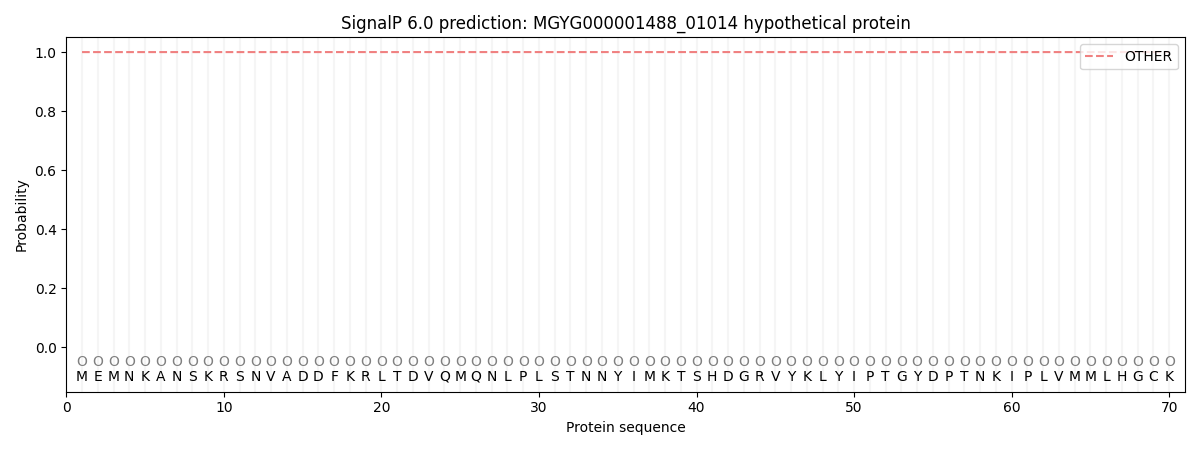
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000073 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |