You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001507_00796
You are here: Home > Sequence: MGYG000001507_00796
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus ihuae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus ihuae | |||||||||||
| CAZyme ID | MGYG000001507_00796 | |||||||||||
| CAZy Family | GH66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 835953; End: 841064 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH66 | 45 | 442 | 7.7e-111 | 0.6438848920863309 |
| CBM35 | 580 | 689 | 5.7e-22 | 0.9495798319327731 |
| CBM35 | 457 | 571 | 5.5e-18 | 0.907563025210084 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14745 | GH66 | 4.78e-90 | 143 | 454 | 1 | 275 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
| pfam13199 | Glyco_hydro_66 | 1.64e-85 | 49 | 452 | 3 | 368 | Glycosyl hydrolase family 66. This family is a set of glycosyl hydrolase enzymes including cycloisomaltooligosaccharide glucanotransferase (EC:2.4.1.-) and dextranase (EC:3.2.1.11) activities. |
| pfam13199 | Glyco_hydro_66 | 1.46e-39 | 1070 | 1276 | 358 | 557 | Glycosyl hydrolase family 66. This family is a set of glycosyl hydrolase enzymes including cycloisomaltooligosaccharide glucanotransferase (EC:2.4.1.-) and dextranase (EC:3.2.1.11) activities. |
| cd14745 | GH66 | 1.09e-18 | 1070 | 1141 | 266 | 331 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
| NF033190 | inl_like_NEAT_1 | 2.10e-15 | 1528 | 1696 | 582 | 746 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSF46225.1 | 0.0 | 1 | 1703 | 1 | 1703 |
| AIQ39879.1 | 0.0 | 1 | 1703 | 1 | 1701 |
| AEX15594.1 | 0.0 | 29 | 1702 | 38 | 1692 |
| AIQ34445.1 | 0.0 | 39 | 1702 | 46 | 1701 |
| BCG57719.1 | 0.0 | 1 | 1702 | 1 | 1980 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WNK_A | 5.98e-84 | 42 | 569 | 27 | 526 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNP_A | 2.12e-83 | 42 | 569 | 8 | 507 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNP_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNL_A | 5.25e-83 | 42 | 569 | 8 | 507 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNM_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 5X7G_A | 2.48e-78 | 44 | 569 | 28 | 525 | CrystalStructure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase [Paenibacillus sp. 598K],5X7H_A Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose [Paenibacillus sp. 598K] |
| 5AXG_A | 2.10e-46 | 49 | 451 | 51 | 419 | Crystalstructure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223],5AXG_B Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94286 | 2.17e-84 | 3 | 937 | 9 | 870 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 PE=1 SV=1 |
| P70873 | 1.58e-77 | 4 | 937 | 2 | 862 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 GN=cit PE=3 SV=1 |
| P38536 | 1.57e-35 | 1324 | 1703 | 1502 | 1856 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P19424 | 1.19e-25 | 1528 | 1702 | 41 | 214 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| C6CRV0 | 2.25e-21 | 1499 | 1695 | 1254 | 1452 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
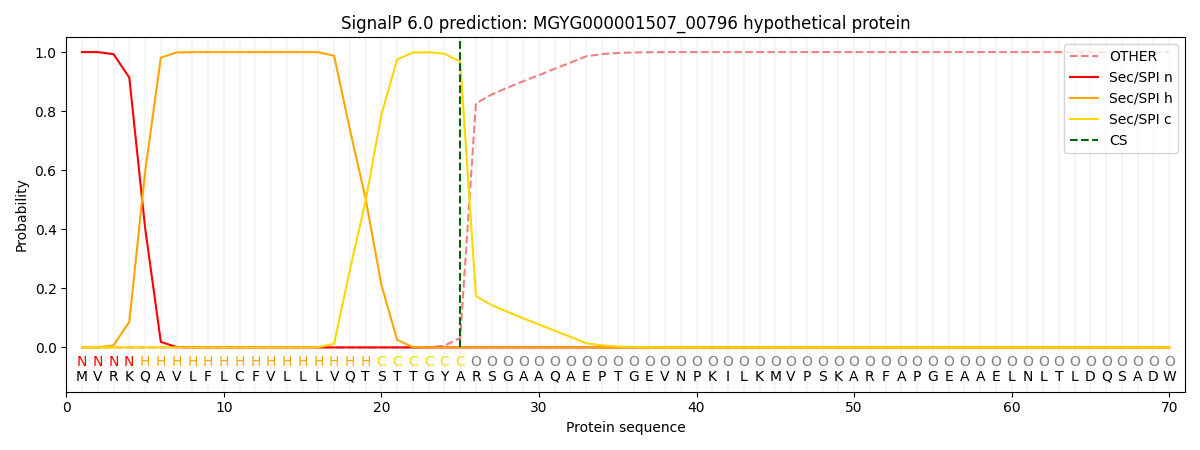
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000235 | 0.999159 | 0.000145 | 0.000169 | 0.000141 | 0.000135 |
