You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001535_03877
You are here: Home > Sequence: MGYG000001535_03877
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A phocaensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A phocaensis | |||||||||||
| CAZyme ID | MGYG000001535_03877 | |||||||||||
| CAZy Family | CBM6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 103190; End: 104608 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00030 | Crystall | 4.07e-11 | 348 | 426 | 1 | 80 | Beta/Gamma crystallin. The alignment comprises two Greek key motifs since the similarity between them is very low. |
| smart00247 | XTALbg | 6.44e-10 | 348 | 426 | 1 | 80 | Beta/gamma crystallins. Beta/gamma crystallins |
| smart00247 | XTALbg | 4.17e-05 | 388 | 443 | 3 | 53 | Beta/gamma crystallins. Beta/gamma crystallins |
| smart00247 | XTALbg | 8.86e-05 | 432 | 470 | 1 | 40 | Beta/gamma crystallins. Beta/gamma crystallins |
| pfam00030 | Crystall | 1.24e-04 | 388 | 443 | 3 | 53 | Beta/Gamma crystallin. The alignment comprises two Greek key motifs since the similarity between them is very low. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARU27134.1 | 5.05e-129 | 25 | 384 | 27 | 386 |
| AYM53477.1 | 5.76e-128 | 18 | 435 | 12 | 455 |
| AYM53674.1 | 5.76e-128 | 18 | 435 | 12 | 455 |
| AYM53558.1 | 3.77e-123 | 18 | 385 | 12 | 383 |
| AYM53603.1 | 3.77e-123 | 18 | 385 | 12 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1PRR_A | 5.01e-25 | 304 | 468 | 6 | 172 | ChainA, DEVELOPMENT-SPECIFIC PROTEIN S [Myxococcus xanthus],1PRS_A Chain A, DEVELOPMENT-SPECIFIC PROTEIN S [Myxococcus xanthus] |
| 3SO1_A | 1.52e-20 | 390 | 471 | 9 | 90 | Crystalstructure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_B Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_C Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_D Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_E Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_F Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_G Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_H Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3SO0_A | 3.91e-20 | 390 | 471 | 9 | 90 | Crystalstructure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_B Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_C Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_D Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_E Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_F Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_G Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_H Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3SNZ_A | 3.91e-20 | 390 | 471 | 9 | 90 | Crystalstructure of a mutant W39D of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3IAJ_A | 7.64e-20 | 390 | 471 | 6 | 87 | Crystalstructure of a betagamma-crystallin domain from Clostridium beijerinckii-in alternate space group I422 [Clostridium beijerinckii NCIMB 8052] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P02966 | 2.74e-24 | 304 | 468 | 6 | 172 | Development-specific protein S OS=Myxococcus xanthus OX=34 GN=tps PE=1 SV=1 |
| P02967 | 2.69e-23 | 304 | 468 | 6 | 173 | Development-specific protein S homolog OS=Myxococcus xanthus OX=34 GN=ops PE=3 SV=1 |
| P46058 | 2.93e-11 | 349 | 470 | 4 | 126 | Epidermal differentiation-specific protein OS=Cynops pyrrhogaster OX=8330 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
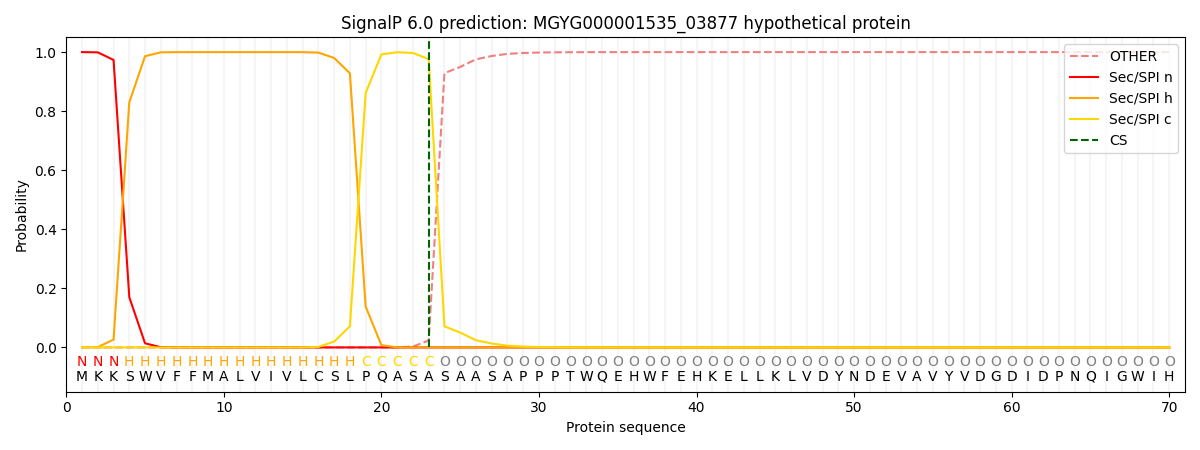
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000231 | 0.999076 | 0.000232 | 0.000162 | 0.000149 | 0.000139 |
