You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_03487
You are here: Home > Sequence: MGYG000001537_03487
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
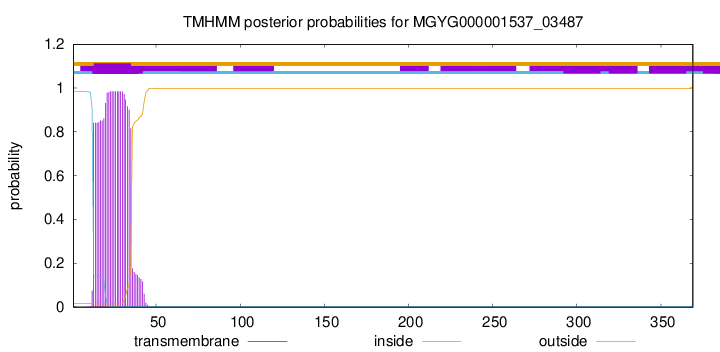
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_03487 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 827429; End: 828538 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 41 | 336 | 7.5e-111 | 0.9834983498349835 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.22e-146 | 40 | 336 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 6.74e-123 | 82 | 334 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.17e-81 | 22 | 331 | 7 | 334 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCB93557.1 | 1.43e-225 | 1 | 346 | 1 | 346 |
| QHT57581.1 | 3.64e-225 | 1 | 346 | 1 | 346 |
| VEH25946.1 | 1.69e-224 | 1 | 346 | 1 | 345 |
| AEE44238.1 | 1.69e-224 | 1 | 346 | 1 | 345 |
| QVI64494.1 | 1.17e-223 | 1 | 346 | 1 | 345 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M0K_A | 2.06e-166 | 1 | 342 | 1 | 339 | CRYSTALSTRUCTURE of endo-1,4-beta-xylanase from Cellulomonas flavigena [Cellulomonas flavigena],5M0K_B CRYSTAL STRUCTURE of endo-1,4-beta-xylanase from Cellulomonas flavigena [Cellulomonas flavigena] |
| 1ISV_A | 2.08e-165 | 36 | 360 | 1 | 317 | Crystalstructure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose [Streptomyces olivaceoviridis],1ISV_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose [Streptomyces olivaceoviridis],1ISW_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose [Streptomyces olivaceoviridis],1ISW_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose [Streptomyces olivaceoviridis],1ISX_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose [Streptomyces olivaceoviridis],1ISX_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose [Streptomyces olivaceoviridis],1ISY_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose [Streptomyces olivaceoviridis],1ISY_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose [Streptomyces olivaceoviridis],1ISZ_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose [Streptomyces olivaceoviridis],1ISZ_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose [Streptomyces olivaceoviridis],1IT0_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose [Streptomyces olivaceoviridis],1IT0_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose [Streptomyces olivaceoviridis],1V6U_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose [Streptomyces olivaceoviridis],1V6U_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose [Streptomyces olivaceoviridis],1V6V_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose [Streptomyces olivaceoviridis],1V6V_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose [Streptomyces olivaceoviridis],1V6W_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose [Streptomyces olivaceoviridis],1V6W_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose [Streptomyces olivaceoviridis],1V6X_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose [Streptomyces olivaceoviridis],1V6X_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose [Streptomyces olivaceoviridis],1XYF_A Endo-1,4-Beta-Xylanase From Streptomyces Olivaceoviridis [Streptomyces olivaceoviridis],1XYF_B Endo-1,4-Beta-Xylanase From Streptomyces Olivaceoviridis [Streptomyces olivaceoviridis] |
| 2G3I_A | 1.48e-164 | 36 | 341 | 1 | 303 | Structureof S.olivaceoviridis xylanase Q88A/R275A mutant [Streptomyces olivaceoviridis],2G3J_A Structure of S.olivaceoviridis xylanase Q88A/R275A mutant [Streptomyces olivaceoviridis],2G4F_A Chain A, Hydrolase [Streptomyces olivaceoviridis],2G4F_B Chain B, Hydrolase [Streptomyces olivaceoviridis] |
| 5GQD_A | 1.69e-164 | 36 | 360 | 1 | 317 | Crystalstructure of covalent glycosyl-enzyme intermediate of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],5GQD_B Crystal structure of covalent glycosyl-enzyme intermediate of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],5GQE_A Crystal structure of michaelis complex of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],5GQE_B Crystal structure of michaelis complex of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis] |
| 2D1Z_A | 6.85e-164 | 36 | 360 | 1 | 317 | Crystalstructure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D1Z_B Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D20_A Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D20_B Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D22_A Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D22_B Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D23_A Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D23_B Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D24_A Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],2D24_B Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26514 | 1.75e-161 | 35 | 360 | 41 | 358 | Endo-1,4-beta-xylanase A OS=Streptomyces lividans OX=1916 GN=xlnA PE=1 SV=2 |
| P07986 | 6.45e-106 | 2 | 346 | 4 | 361 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
| P23360 | 2.66e-86 | 50 | 338 | 43 | 327 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
| Q0H904 | 4.25e-84 | 36 | 338 | 27 | 325 | Endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnC PE=2 SV=2 |
| B0Y6E0 | 5.80e-82 | 36 | 338 | 27 | 316 | Probable endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xlnC PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
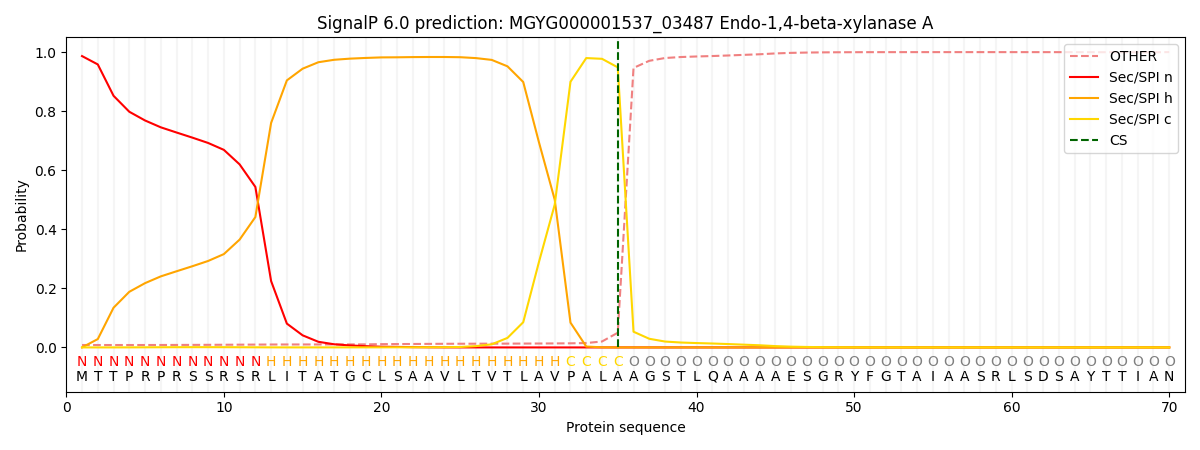
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.009349 | 0.983904 | 0.003748 | 0.002503 | 0.000271 | 0.000198 |