You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001546_01304
You are here: Home > Sequence: MGYG000001546_01304
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes provencensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes provencensis | |||||||||||
| CAZyme ID | MGYG000001546_01304 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1616934; End: 1619756 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 18 | 505 | 3.1e-64 | 0.4920212765957447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.01e-19 | 21 | 788 | 10 | 788 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 5.97e-18 | 21 | 472 | 10 | 440 | beta-D-glucuronidase; Provisional |
| pfam02836 | Glyco_hydro_2_C | 7.92e-07 | 330 | 477 | 1 | 159 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02837 | Glyco_hydro_2_N | 5.89e-05 | 26 | 219 | 4 | 169 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 7.48e-04 | 223 | 328 | 3 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYB35766.1 | 0.0 | 24 | 937 | 50 | 968 |
| BBE19496.1 | 0.0 | 19 | 936 | 22 | 941 |
| QEL03576.1 | 0.0 | 1 | 936 | 3 | 941 |
| AQT69150.1 | 0.0 | 19 | 937 | 20 | 939 |
| QTO24394.1 | 0.0 | 3 | 936 | 4 | 924 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 4.34e-18 | 24 | 451 | 39 | 440 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 4.34e-18 | 24 | 451 | 40 | 441 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 1YQ2_A | 4.36e-16 | 18 | 451 | 28 | 443 | ChainA, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_B Chain B, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_C Chain C, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_D Chain D, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_E Chain E, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_F Chain F, beta-galactosidase [Arthrobacter sp. C2-2] |
| 6D8K_A | 5.21e-11 | 21 | 412 | 33 | 374 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
| 7VQM_A | 3.62e-10 | 21 | 462 | 11 | 421 | ChainA, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 2.37e-17 | 24 | 451 | 40 | 441 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O77695 | 2.88e-15 | 12 | 472 | 24 | 471 | Beta-glucuronidase (Fragment) OS=Chlorocebus aethiops OX=9534 GN=GUSB PE=2 SV=1 |
| Q2XQU3 | 1.23e-14 | 19 | 451 | 49 | 463 | Beta-galactosidase 2 OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
| Q5R5N6 | 2.00e-14 | 12 | 472 | 27 | 474 | Beta-glucuronidase OS=Pongo abelii OX=9601 GN=GUSB PE=2 SV=2 |
| Q6LL68 | 4.82e-14 | 16 | 451 | 41 | 456 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
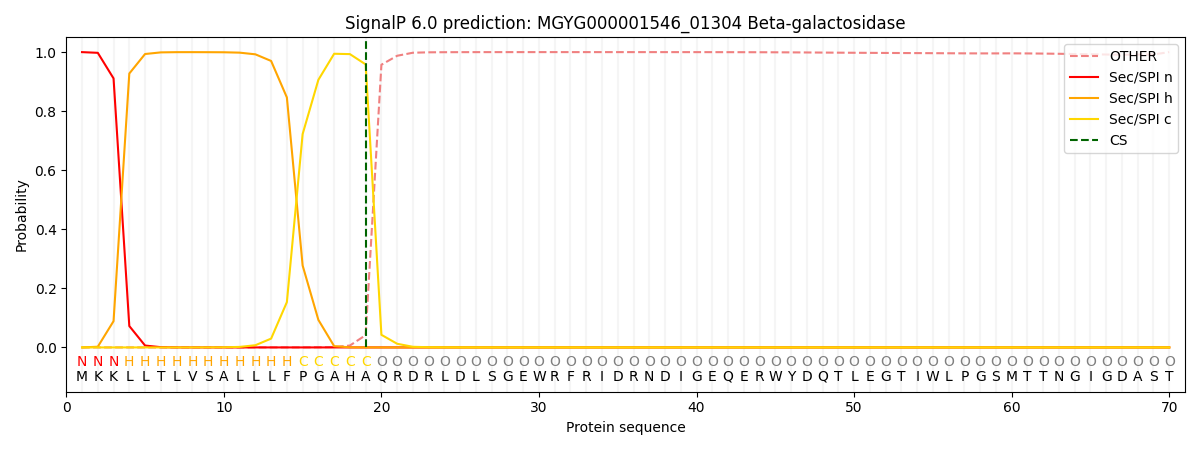
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005274 | 0.985799 | 0.005180 | 0.001646 | 0.000887 | 0.001179 |
