You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001562_02405
You are here: Home > Sequence: MGYG000001562_02405
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes timonensis | |||||||||||
| CAZyme ID | MGYG000001562_02405 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24986; End: 26533 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 19 | 371 | 2.7e-49 | 0.8688946015424165 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03901 | Glyco_transf_22 | 7.62e-11 | 14 | 319 | 1 | 312 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| PLN02816 | PLN02816 | 1.89e-10 | 8 | 319 | 34 | 339 | mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL06999.1 | 1.60e-202 | 3 | 513 | 8 | 515 |
| AUR51123.1 | 4.65e-61 | 12 | 504 | 9 | 472 |
| AOW16884.1 | 6.95e-57 | 20 | 340 | 17 | 331 |
| QVY67119.1 | 5.56e-56 | 24 | 346 | 21 | 338 |
| QHS61266.1 | 4.06e-49 | 15 | 340 | 8 | 325 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q94A15 | 3.18e-19 | 8 | 327 | 34 | 347 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
| Q9JJQ0 | 1.81e-11 | 35 | 387 | 73 | 411 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
| Q1LZA0 | 2.39e-11 | 35 | 366 | 74 | 396 | GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1 |
| Q92521 | 1.63e-09 | 35 | 327 | 84 | 367 | GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1 |
| Q7SXZ1 | 3.21e-07 | 3 | 387 | 34 | 404 | GPI mannosyltransferase 3 OS=Danio rerio OX=7955 GN=pigb PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
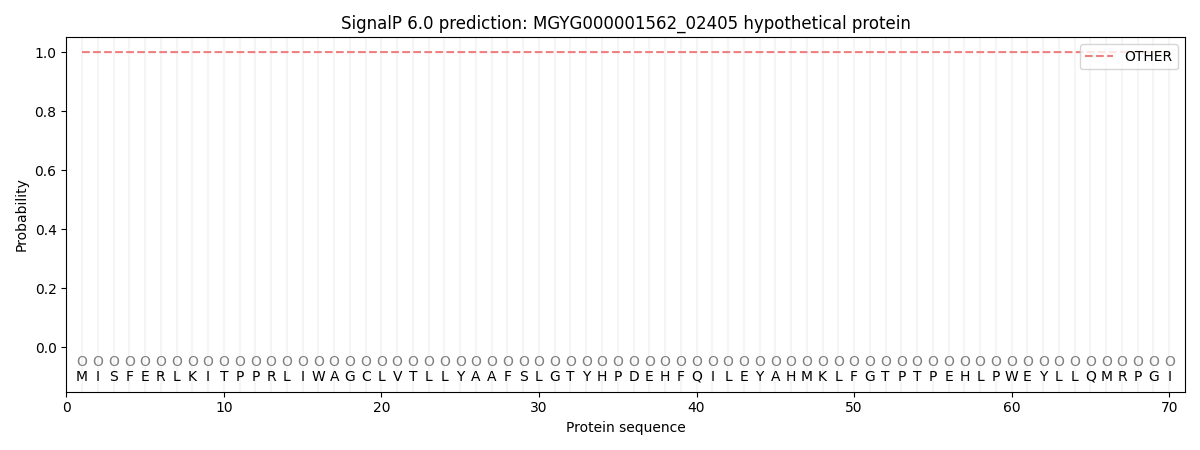
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000042 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
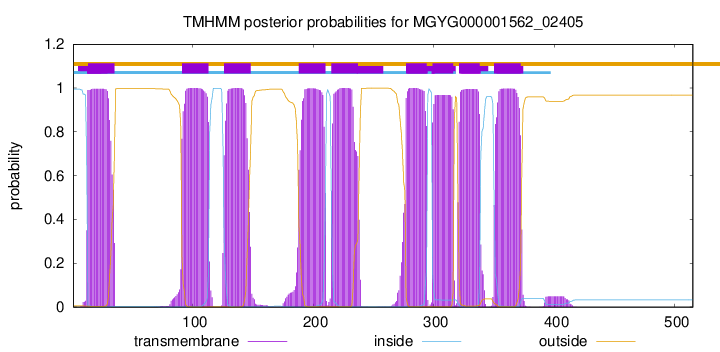
| start | end |
|---|---|
| 13 | 35 |
| 91 | 113 |
| 126 | 148 |
| 188 | 210 |
| 215 | 237 |
| 277 | 294 |
| 299 | 316 |
| 321 | 338 |
| 350 | 372 |
