You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001652_00086
You are here: Home > Sequence: MGYG000001652_00086
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
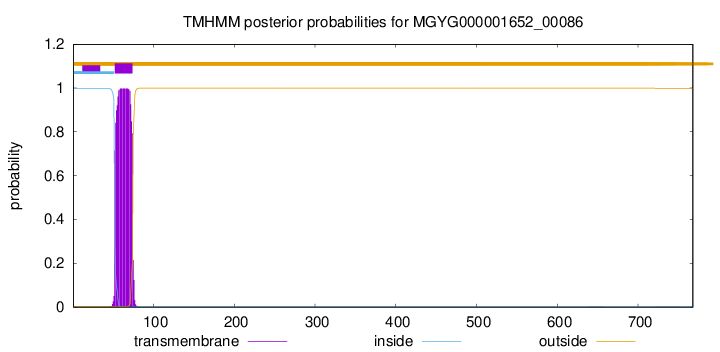
TMHMM annotations
Basic Information help
| Species | V9D3004 sp002349525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; V9D3004; V9D3004 sp002349525 | |||||||||||
| CAZyme ID | MGYG000001652_00086 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 114301; End: 116607 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 6.92e-22 | 427 | 658 | 45 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 9.11e-09 | 90 | 152 | 1 | 54 | Bacterial SH3 domain. |
| COG3103 | YgiM | 4.68e-06 | 64 | 253 | 7 | 166 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| cd11856 | SH3_p47phox_like | 0.002 | 114 | 153 | 18 | 53 | Src homology 3 domains of the p47phox subunit of NADPH oxidase and similar domains. This family is composed of the tandem SH3 domains of p47phox subunit of NADPH oxidase and Nox Organizing protein 1 (NoxO1), the four SH3 domains of Tks4 (Tyr kinase substrate with four SH3 domains), the five SH3 domains of Tks5, the SH3 domain of obscurin, Myosin-I, and similar domains. Most members of this group also contain Phox homology (PX) domains, except for obscurin and Myosin-I. p47phox and NoxO1 are regulators of the phagocytic NADPH oxidase complex (also called Nox2 or gp91phox) and nonphagocytic NADPH oxidase Nox1, respectively. They play roles in the activation of their respective NADPH oxidase, which catalyzes the transfer of electrons from NADPH to molecular oxygen to form superoxide. Tks proteins are Src substrates and scaffolding proteins that play important roles in the formation of podosomes and invadopodia, the dynamic actin-rich structures that are related to cell migration and cancer cell invasion. Obscurin is a giant muscle protein that plays important roles in the organization and assembly of the myofibril and the sarcoplasmic reticulum. Type I myosins (Myosin-I) are actin-dependent motors in endocytic actin structures and actin patches. They play roles in membrane traffic in endocytic and secretory pathways, cell motility, and mechanosensing. Myosin-I contains an N-terminal actin-activated ATPase, a phospholipid-binding TH1 (tail homology 1) domain, and a C-terminal extension which includes an F-actin-binding TH2 domain, an SH3 domain, and an acidic peptide that participates in activating the Arp2/3complex. The SH3 domain of myosin-I is required for myosin-I-induced actin polymerization. SH3 domains are protein interaction domains that bind to proline-rich ligands with moderate affinity and selectivity, preferentially to PxxP motifs. They play versatile and diverse roles in the cell including the regulation of enzymes, changing the subcellular localization of signaling pathway components, and mediating the formation of multiprotein complex assemblies. |
| smart00287 | SH3b | 0.002 | 86 | 148 | 5 | 58 | Bacterial SH3 domain homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHQ61172.1 | 6.70e-148 | 59 | 763 | 13 | 762 |
| BCN29841.1 | 2.86e-140 | 81 | 765 | 31 | 886 |
| BBF45313.1 | 3.29e-139 | 81 | 731 | 31 | 710 |
| BCJ97942.1 | 4.80e-131 | 81 | 765 | 33 | 904 |
| BCJ93456.1 | 1.26e-130 | 76 | 765 | 28 | 892 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 2.86e-09 | 503 | 658 | 95 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CPQ1 | 3.61e-09 | 430 | 658 | 1128 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 8.18e-09 | 430 | 658 | 1128 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 8.18e-09 | 430 | 658 | 1128 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q931U5 | 9.34e-08 | 503 | 658 | 1099 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| Q99V41 | 9.34e-08 | 503 | 658 | 1099 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
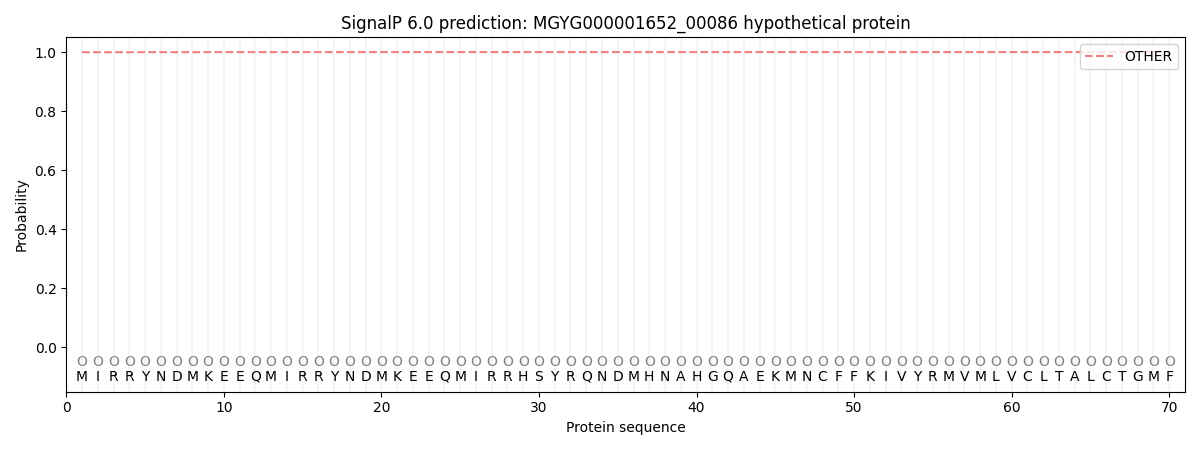
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999634 | 0.000315 | 0.000010 | 0.000000 | 0.000000 | 0.000054 |