You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001666_00144
You are here: Home > Sequence: MGYG000001666_00144
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; | |||||||||||
| CAZyme ID | MGYG000001666_00144 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57; End: 2141 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 434 | 689 | 8.7e-48 | 0.6798679867986799 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 3.11e-36 | 434 | 687 | 54 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 7.22e-36 | 434 | 689 | 96 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.12e-18 | 438 | 689 | 123 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 1.77e-09 | 76 | 140 | 35 | 98 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.24e-07 | 76 | 153 | 58 | 129 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD43725.1 | 1.81e-286 | 169 | 694 | 1 | 527 |
| ALJ61518.1 | 3.05e-272 | 11 | 694 | 23 | 721 |
| QRQ48288.1 | 8.96e-215 | 9 | 694 | 20 | 719 |
| QUT46089.1 | 8.22e-213 | 9 | 694 | 20 | 719 |
| ADE81777.1 | 1.64e-209 | 8 | 694 | 18 | 707 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1US3_A | 7.23e-16 | 455 | 691 | 291 | 513 | Nativexylanase10C from Cellvibrio japonicus [Cellvibrio japonicus] |
| 4XUY_A | 7.81e-16 | 433 | 691 | 101 | 302 | Crystalstructure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
| 1K6A_A | 1.06e-15 | 442 | 691 | 108 | 301 | Structuralstudies on the mobility in the active site of the Thermoascus aurantiacus xylanase I [Thermoascus aurantiacus] |
| 1GOK_A | 1.06e-15 | 442 | 691 | 108 | 301 | Thermostablexylanase I from Thermoascus aurantiacus- Crystal form II [Thermoascus aurantiacus],1GOM_A Thermostable xylanase I from Thermoascus aurantiacus- Crystal form I [Thermoascus aurantiacus],1GOO_A Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex [Thermoascus aurantiacus],1GOQ_A Thermostable xylanase I from Thermoascus aurantiacus - Room temperature xylobiose complex [Thermoascus aurantiacus],1GOR_A THERMOSTABLE XYLANASE I FROM THERMOASCUS AURANTIACUS - XYLOBIOSE COMPLEX AT 100 K [Thermoascus aurantiacus] |
| 5Y3X_A | 1.97e-15 | 437 | 671 | 131 | 331 | Crystalstructure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_B Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_C Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_D Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_E Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_F Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4P902 | 3.55e-17 | 437 | 693 | 121 | 341 | Endo-1,4-beta-xylanase UM03411 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_03411 PE=1 SV=1 |
| P33559 | 4.36e-15 | 433 | 691 | 126 | 327 | Endo-1,4-beta-xylanase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xynA PE=1 SV=2 |
| Q59675 | 4.80e-15 | 455 | 691 | 375 | 597 | Endo-beta-1,4-xylanase Xyn10C OS=Cellvibrio japonicus OX=155077 GN=xyn10C PE=1 SV=2 |
| A2QFV7 | 5.85e-15 | 433 | 691 | 126 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnC PE=1 SV=1 |
| O59859 | 7.85e-15 | 442 | 691 | 134 | 327 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
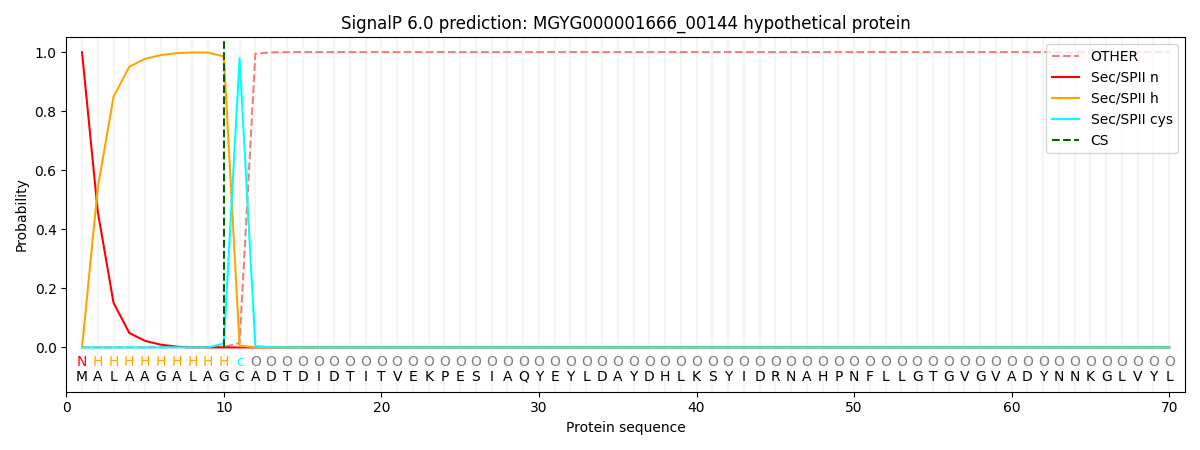
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000162 | 0.000228 | 0.999647 | 0.000000 | 0.000001 | 0.000000 |
