You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001686_00411
You are here: Home > Sequence: MGYG000001686_00411
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | DNF00809 sp900604965 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Eggerthellaceae; DNF00809; DNF00809 sp900604965 | |||||||||||
| CAZyme ID | MGYG000001686_00411 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1864; End: 3951 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 8.87e-45 | 536 | 694 | 93 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 2.65e-20 | 537 | 679 | 7 | 143 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| NF033435 | S-layer_Clost | 2.91e-16 | 121 | 348 | 380 | 617 | S-layer protein SlpA. In Clostridiodes difficile, the S-layer protein precursor, SlpA, is one member of a large paralogous family of protein that share several cell wall-binding repeats. SlpA is cleaved into a larger and smaller protein. The S-layer protein itself is important to adhesion, and portions of it are highly variable, and then N-terminal and C-terminal are well-conserved. |
| pfam01832 | Glucosaminidase | 3.68e-11 | 543 | 629 | 1 | 78 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| pfam04122 | CW_binding_2 | 9.81e-11 | 339 | 428 | 1 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKF06896.1 | 1.76e-173 | 10 | 694 | 11 | 697 |
| QOS67179.1 | 3.06e-110 | 125 | 695 | 161 | 715 |
| BCA89043.1 | 1.13e-42 | 454 | 694 | 797 | 1047 |
| BCS56855.1 | 2.79e-42 | 490 | 694 | 859 | 1071 |
| BAE05645.1 | 2.22e-29 | 460 | 680 | 429 | 636 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PI7_A | 2.56e-22 | 536 | 684 | 77 | 221 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 1.60e-21 | 536 | 684 | 77 | 221 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXO_A | 3.07e-21 | 519 | 663 | 72 | 216 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXP_A | 5.12e-21 | 536 | 677 | 94 | 232 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 1.14e-20 | 536 | 677 | 124 | 262 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 3.79e-25 | 536 | 694 | 728 | 879 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q99V41 | 1.18e-18 | 519 | 663 | 1076 | 1220 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q931U5 | 1.18e-18 | 519 | 663 | 1076 | 1220 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| Q6GI31 | 1.19e-18 | 519 | 663 | 1085 | 1229 | Bifunctional autolysin OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=atl PE=3 SV=1 |
| Q2FZK7 | 1.19e-18 | 519 | 663 | 1084 | 1228 | Bifunctional autolysin OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
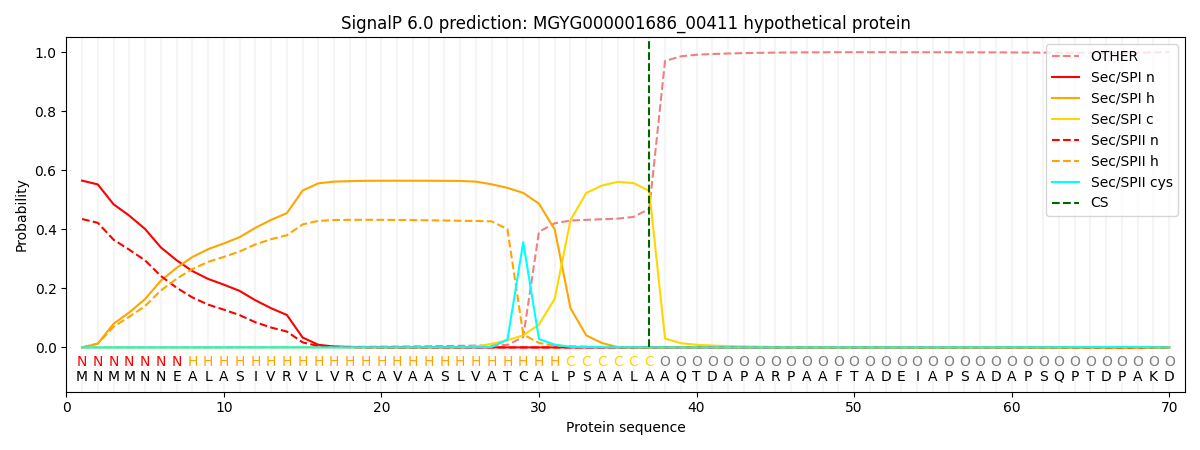
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000709 | 0.554752 | 0.443295 | 0.000706 | 0.000327 | 0.000198 |
