You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001691_00924
You are here: Home > Sequence: MGYG000001691_00924
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aeromonas veronii_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Aeromonadaceae; Aeromonas; Aeromonas veronii_A | |||||||||||
| CAZyme ID | MGYG000001691_00924 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | GlcNAc-binding protein A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11567; End: 13756 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 36 | 285 | 1.3e-29 | 0.6756756756756757 |
| CBM73 | 677 | 727 | 1.3e-20 | 0.9444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3469 | Chi1 | 1.10e-148 | 22 | 343 | 13 | 332 | Chitinase [Carbohydrate transport and metabolism]. |
| cd02871 | GH18_chitinase_D-like | 1.19e-103 | 36 | 339 | 2 | 312 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| PRK13211 | PRK13211 | 1.10e-27 | 567 | 729 | 306 | 477 | N-acetylglucosamine-binding protein GbpA. |
| pfam00704 | Glyco_hydro_18 | 1.89e-19 | 36 | 318 | 1 | 307 | Glycosyl hydrolases family 18. |
| pfam16403 | DUF5011 | 4.99e-19 | 434 | 503 | 4 | 71 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSR71778.1 | 0.0 | 1 | 729 | 1 | 730 |
| QQB21890.1 | 0.0 | 1 | 729 | 1 | 730 |
| QWL59697.1 | 0.0 | 1 | 729 | 1 | 730 |
| QWL67876.1 | 0.0 | 1 | 729 | 1 | 730 |
| QTL96393.1 | 0.0 | 1 | 729 | 1 | 730 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HMC_A | 3.20e-243 | 36 | 554 | 10 | 528 | Crystalstructure of cold-adapted chitinase from Moritella marina [Moritella marina],4HMD_A Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) [Moritella marina],4HME_A Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 [Moritella marina] |
| 4MB3_A | 9.11e-243 | 36 | 554 | 10 | 528 | Crystalstructure of E153Q mutant of cold-adapted chitinase from Moritella marina [Moritella marina],4MB4_A Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 [Moritella marina],4MB5_A Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 [Moritella marina] |
| 4W5Z_A | 5.29e-173 | 36 | 343 | 32 | 341 | Highresolution crystal structure of catalytic domain of Chitinase 60 from psychrophilic bacteria Moritella marina. [Moritella marina] |
| 4AXN_A | 9.86e-126 | 36 | 340 | 26 | 327 | Hallmarksof processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases [Serratia marcescens],4AXN_B Hallmarks of processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases [Serratia marcescens] |
| 3IAN_A | 3.36e-121 | 36 | 340 | 6 | 310 | ChainA, Chitinase [Lactococcus lactis subsp. lactis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q838S2 | 1.22e-126 | 18 | 340 | 24 | 347 | Chitinase OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=EF_0361 PE=1 SV=1 |
| P27050 | 8.34e-31 | 37 | 350 | 192 | 521 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| Q8NJQ5 | 6.38e-21 | 20 | 322 | 23 | 331 | Endochitinase 37 OS=Trichoderma harzianum OX=5544 GN=chit37 PE=1 SV=1 |
| E9F7R6 | 4.28e-20 | 32 | 318 | 38 | 330 | Endochitinase 4 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chi4 PE=3 SV=1 |
| A5FB63 | 9.29e-20 | 34 | 343 | 1141 | 1483 | Chitinase ChiA OS=Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / NBRC 14942 / NCIMB 11054 / UW101) OX=376686 GN=chiA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
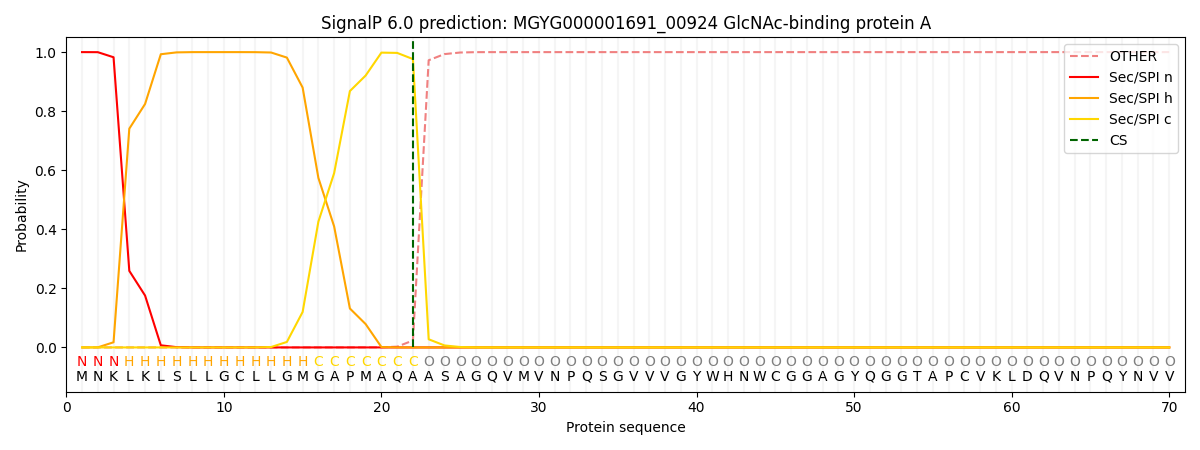
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000281 | 0.998985 | 0.000200 | 0.000180 | 0.000166 | 0.000147 |
