You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001770_02389
You are here: Home > Sequence: MGYG000001770_02389
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900313215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900313215 | |||||||||||
| CAZyme ID | MGYG000001770_02389 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2424; End: 3773 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 22 | 210 | 7.8e-44 | 0.9845360824742269 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01833 | XynB_like | 1.61e-26 | 20 | 210 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| COG0657 | Aes | 1.65e-20 | 193 | 431 | 21 | 298 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
| COG1506 | DAP2 | 2.23e-16 | 221 | 419 | 363 | 594 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam07859 | Abhydrolase_3 | 5.65e-15 | 253 | 419 | 1 | 205 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| pfam13472 | Lipase_GDSL_2 | 1.40e-13 | 25 | 198 | 2 | 173 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAX78799.1 | 6.07e-133 | 16 | 449 | 34 | 494 |
| ATC65549.1 | 1.80e-107 | 10 | 431 | 28 | 477 |
| QOV88633.1 | 1.03e-63 | 18 | 214 | 28 | 226 |
| QNN20991.1 | 3.16e-58 | 16 | 214 | 26 | 226 |
| QDT00894.1 | 3.68e-31 | 226 | 449 | 36 | 283 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AO9_A | 1.45e-28 | 208 | 449 | 1 | 278 | Thestructure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-native [Thermogutta terrifontis],5AOA_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-Propionate bound [Thermogutta terrifontis],5AOB_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-butyrate bound [Thermogutta terrifontis],5AOC_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-valerate bound [Thermogutta terrifontis] |
| 7BFN_A | 1.48e-28 | 208 | 449 | 2 | 279 | ChainA, Esterase [Thermogutta terrifontis] |
| 7BFO_A | 7.37e-28 | 208 | 449 | 2 | 279 | ChainA, Esterase [Thermogutta terrifontis],7BFR_A Chain A, Esterase [Thermogutta terrifontis],7BFT_A Chain A, Esterase [Thermogutta terrifontis],7BFU_A Chain A, Esterase [Thermogutta terrifontis],7BFV_A Chain A, Esterase [Thermogutta terrifontis] |
| 2C7B_A | 7.55e-11 | 233 | 441 | 56 | 302 | TheCrystal Structure of EstE1, a New Thermophilic and Thermostable Carboxylesterase Cloned from a Metagenomic Library [uncultured archaeon],2C7B_B The Crystal Structure of EstE1, a New Thermophilic and Thermostable Carboxylesterase Cloned from a Metagenomic Library [uncultured archaeon] |
| 1QZ3_A | 5.77e-10 | 234 | 402 | 57 | 222 | CRYSTALSTRUCTURE OF MUTANT M211S/R215L OF CARBOXYLESTERASE EST2 COMPLEXED WITH HEXADECANESULFONATE [Alicyclobacillus acidocaldarius],1U4N_A Crystal Structure Analysis of the M211S/R215L EST2 mutant [Alicyclobacillus acidocaldarius] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9US38 | 2.14e-07 | 232 | 359 | 81 | 206 | AB hydrolase superfamily protein C1039.03 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC1039.03 PE=3 SV=1 |
| D2D3B6 | 3.33e-07 | 233 | 375 | 126 | 279 | Fumonisin B1 esterase OS=Sphingopyxis macrogoltabida OX=33050 GN=fumD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
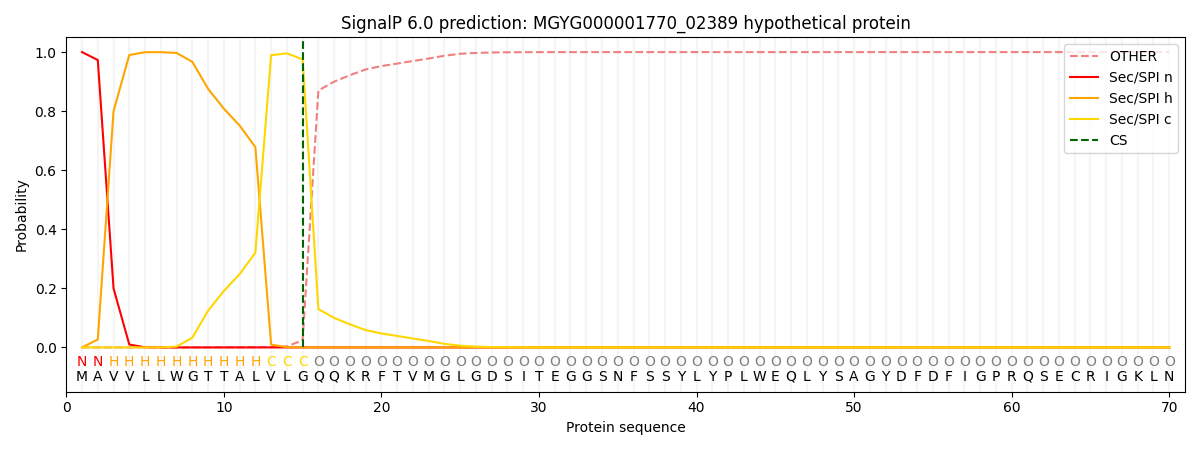
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000346 | 0.999036 | 0.000154 | 0.000165 | 0.000145 | 0.000145 |
