You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001774_01638
You are here: Home > Sequence: MGYG000001774_01638
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-831 sp000432775 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; CAG-831; CAG-831 sp000432775 | |||||||||||
| CAZyme ID | MGYG000001774_01638 | |||||||||||
| CAZy Family | CE19 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27861; End: 29012 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE19 | 78 | 381 | 4.1e-28 | 0.7771084337349398 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0412 | DLH | 2.11e-16 | 110 | 271 | 13 | 141 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam12715 | Abhydrolase_7 | 2.96e-15 | 56 | 342 | 45 | 327 | Abhydrolase family. This is a family of probable bacterial abhydrolases. |
| COG1506 | DAP2 | 5.69e-12 | 91 | 383 | 358 | 617 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam01738 | DLH | 2.63e-06 | 112 | 352 | 1 | 173 | Dienelactone hydrolase family. |
| COG1073 | FrsA | 1.43e-05 | 97 | 346 | 19 | 261 | Fermentation-respiration switch protein FrsA, has esterase activity, DUF1100 family [Signal transduction mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37157.1 | 7.95e-162 | 27 | 381 | 373 | 726 |
| AMV40306.1 | 2.28e-22 | 51 | 367 | 47 | 346 |
| QEH40625.1 | 3.91e-22 | 52 | 374 | 38 | 349 |
| QDV32533.1 | 7.77e-21 | 51 | 383 | 29 | 342 |
| APW62647.1 | 6.79e-20 | 33 | 383 | 40 | 373 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3G8Y_A | 8.18e-12 | 56 | 340 | 46 | 326 | ChainA, SusD/RagB-associated esterase-like protein [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34973 | 3.71e-26 | 91 | 383 | 30 | 299 | Putative hydrolase YtaP OS=Bacillus subtilis (strain 168) OX=224308 GN=ytaP PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
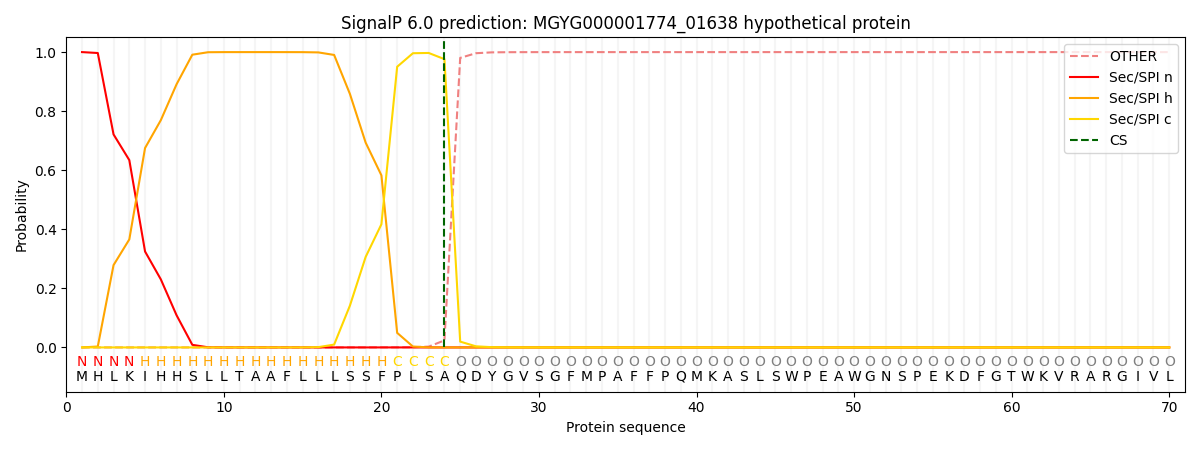
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000373 | 0.998869 | 0.000208 | 0.000202 | 0.000184 | 0.000169 |
