You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001781_01130
You are here: Home > Sequence: MGYG000001781_01130
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acutalibacter sp000432995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Acutalibacter; Acutalibacter sp000432995 | |||||||||||
| CAZyme ID | MGYG000001781_01130 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22574; End: 23773 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 44 | 382 | 8.6e-98 | 0.9933993399339934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 5.53e-104 | 45 | 382 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 5.93e-95 | 88 | 378 | 2 | 261 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.03e-84 | 54 | 378 | 38 | 335 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEE95681.1 | 7.28e-95 | 40 | 380 | 47 | 375 |
| BCZ47833.1 | 3.02e-90 | 38 | 387 | 51 | 405 |
| QZN73189.1 | 1.43e-88 | 39 | 386 | 208 | 556 |
| APO44853.1 | 1.99e-88 | 39 | 382 | 208 | 552 |
| BAA34091.1 | 2.07e-87 | 39 | 386 | 209 | 557 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4W8L_A | 5.78e-91 | 43 | 382 | 3 | 343 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 6FHE_A | 5.60e-85 | 38 | 381 | 6 | 339 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 3W24_A | 3.24e-80 | 39 | 383 | 3 | 326 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W25_A | 2.57e-79 | 39 | 383 | 3 | 326 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W26_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W27_A | 2.57e-79 | 39 | 383 | 3 | 326 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W28_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W29_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O69230 | 9.72e-85 | 26 | 382 | 353 | 709 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| O69231 | 6.21e-76 | 43 | 381 | 6 | 329 | Endo-1,4-beta-xylanase B OS=Paenibacillus barcinonensis OX=198119 GN=xynB PE=1 SV=1 |
| P23556 | 9.36e-75 | 44 | 381 | 16 | 340 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=xynA PE=1 SV=1 |
| P36917 | 6.10e-73 | 39 | 397 | 352 | 694 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| P38535 | 2.72e-72 | 39 | 397 | 204 | 546 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
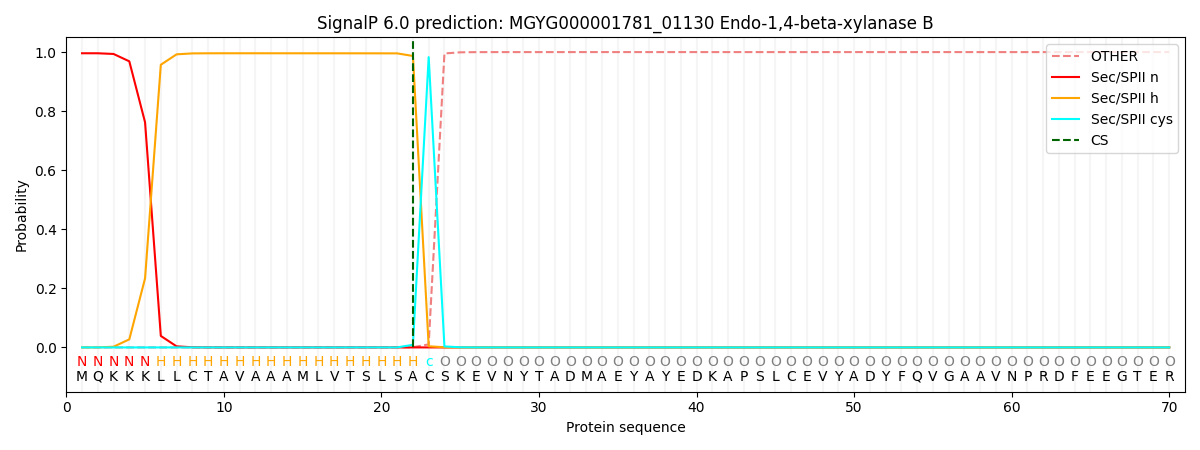
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000004 | 0.003875 | 0.996154 | 0.000001 | 0.000002 | 0.000001 |
