You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001784_02472
Basic Information
help
| Species |
Mailhella sp003150275
|
| Lineage |
Bacteria; Desulfobacterota; Desulfovibrionia; Desulfovibrionales; Desulfovibrionaceae; Mailhella; Mailhella sp003150275
|
| CAZyme ID |
MGYG000001784_02472
|
| CAZy Family |
GT83 |
| CAZyme Description |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001784 |
3348621 |
MAG |
Denmark |
Europe |
|
| Gene Location |
Start: 4928;
End: 6670
Strand: +
|
No EC number prediction in MGYG000001784_02472.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
60 |
468 |
3.6e-48 |
0.7981481481481482 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
1.12e-18 |
86 |
531 |
35 |
502 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
C6DAW3
|
1.21e-14 |
59 |
411 |
8 |
365 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium carotovorum subsp. carotovorum (strain PC1) OX=561230 GN=arnT PE=3 SV=1 |
|
B4ETL9
|
2.00e-13 |
86 |
408 |
36 |
365 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Proteus mirabilis (strain HI4320) OX=529507 GN=arnT2 PE=3 SV=1 |
|
Q7N3Q9
|
1.25e-10 |
86 |
402 |
36 |
359 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Photorhabdus laumondii subsp. laumondii (strain DSM 15139 / CIP 105565 / TT01) OX=243265 GN=arnT PE=2 SV=1 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.998717
|
0.001244
|
0.000024
|
0.000005
|
0.000003
|
0.000014
|
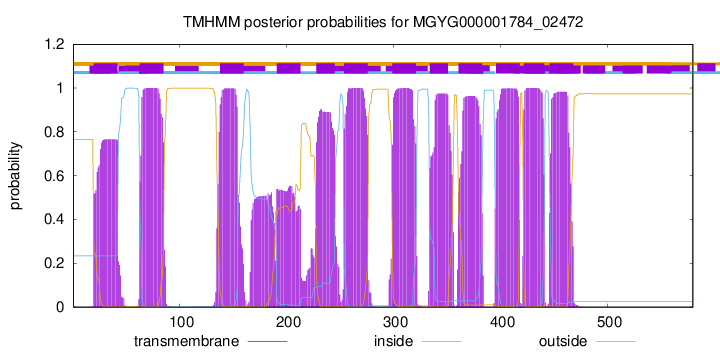
| start |
end |
| 20 |
42 |
| 63 |
85 |
| 138 |
160 |
| 191 |
213 |
| 228 |
245 |
| 254 |
276 |
| 299 |
321 |
| 334 |
351 |
| 361 |
383 |
| 396 |
418 |
| 422 |
441 |
| 446 |
468 |


