You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001803_00655
You are here: Home > Sequence: MGYG000001803_00655
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900546925 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900546925 | |||||||||||
| CAZyme ID | MGYG000001803_00655 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31237; End: 34200 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 48 | 596 | 1.2e-99 | 0.5904255319148937 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd09019 | galactose_mutarotase_like | 1.57e-159 | 654 | 984 | 1 | 326 | galactose mutarotase_like. Galactose mutarotase catalyzes the conversion of beta-D-galactose to alpha-D-galactose. Beta-D-galactose is produced by the degradation of lactose, a disaccharide composed of beta-D-glucose and beta-D-galactose. This epimerization reaction is the first step in the four-step Leloir pathway, which converts galactose into metabolically important glucose. This epimerization step is followed by the phosophorylation of alpha-D-galactose by galactokinase, an enzyme which can only act on the alpha anomer. A glutamate and a histidine residue of the galactose mutarotase have been shown to be critical for catalysis, the glutamate serves as the active site base to initiate the reaction by removing the proton from the C-1 hydroxyl group of the sugar substrate, and the histidine as the active site acid to protonate the C-5 ring oxygen. Galactose mutarotase is a member of the aldose-1-epimerase superfamily. |
| PLN00194 | PLN00194 | 8.92e-120 | 654 | 986 | 11 | 337 | aldose 1-epimerase; Provisional |
| PRK11055 | galM | 2.68e-94 | 653 | 986 | 10 | 341 | galactose-1-epimerase; Provisional |
| pfam01263 | Aldose_epim | 1.16e-82 | 654 | 983 | 2 | 300 | Aldose 1-epimerase. |
| COG2017 | GalM | 1.20e-64 | 649 | 987 | 9 | 308 | Galactose mutarotase or related enzyme [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AST54135.1 | 1.48e-217 | 1 | 625 | 1 | 601 |
| ABR42251.1 | 1.48e-217 | 1 | 625 | 1 | 601 |
| QUT94725.1 | 1.48e-217 | 1 | 625 | 1 | 601 |
| BBK91716.1 | 1.48e-217 | 1 | 625 | 1 | 601 |
| QRO17549.1 | 1.48e-217 | 1 | 625 | 1 | 601 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 2.99e-196 | 18 | 625 | 1 | 585 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 1SNZ_A | 2.17e-74 | 655 | 986 | 24 | 343 | Crystalstructure of apo human galactose mutarotase [Homo sapiens],1SNZ_B Crystal structure of apo human galactose mutarotase [Homo sapiens],1SO0_A Crystal structure of human galactose mutarotase complexed with galactose [Homo sapiens],1SO0_B Crystal structure of human galactose mutarotase complexed with galactose [Homo sapiens],1SO0_C Crystal structure of human galactose mutarotase complexed with galactose [Homo sapiens],1SO0_D Crystal structure of human galactose mutarotase complexed with galactose [Homo sapiens] |
| 4RNL_A | 6.51e-70 | 655 | 987 | 25 | 343 | Thecrystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus [Streptomyces platensis],4RNL_B The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus [Streptomyces platensis],4RNL_C The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus [Streptomyces platensis],4RNL_D The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus [Streptomyces platensis] |
| 1LUR_A | 2.85e-43 | 662 | 985 | 23 | 336 | CrystalStructure of the GalM/aldose Epimerase Homologue from C. elegans, Northeast Structural Genomics Target WR66 [Caenorhabditis elegans],1LUR_B Crystal Structure of the GalM/aldose Epimerase Homologue from C. elegans, Northeast Structural Genomics Target WR66 [Caenorhabditis elegans] |
| 1L7J_A | 1.95e-36 | 653 | 987 | 13 | 339 | ChainA, galactose mutarotase [Lactococcus lactis],1L7J_B Chain B, galactose mutarotase [Lactococcus lactis],1L7K_A Chain A, galactose mutarotase [Lactococcus lactis],1L7K_B Chain B, galactose mutarotase [Lactococcus lactis],1MMU_A Chain A, Aldose 1-epimerase [Lactococcus lactis],1MMU_B Chain B, Aldose 1-epimerase [Lactococcus lactis],1MMX_A Chain A, Aldose 1-epimerase [Lactococcus lactis],1MMX_B Chain B, Aldose 1-epimerase [Lactococcus lactis],1MMY_A Chain A, Aldose 1-epimerase [Lactococcus lactis],1MMY_B Chain B, Aldose 1-epimerase [Lactococcus lactis],1MMZ_A Chain A, Aldose 1-epimerase [Lactococcus lactis],1MMZ_B Chain B, Aldose 1-epimerase [Lactococcus lactis],1MN0_A Chain A, Aldose 1-epimerase [Lactococcus lactis],1MN0_B Chain B, Aldose 1-epimerase [Lactococcus lactis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P05149 | 2.83e-76 | 652 | 987 | 37 | 380 | Aldose 1-epimerase OS=Acinetobacter calcoaceticus OX=471 GN=mro PE=1 SV=1 |
| Q66HG4 | 3.21e-76 | 655 | 986 | 22 | 341 | Galactose mutarotase OS=Rattus norvegicus OX=10116 GN=Galm PE=1 SV=1 |
| Q5EA79 | 6.15e-76 | 655 | 986 | 22 | 341 | Galactose mutarotase OS=Bos taurus OX=9913 GN=GALM PE=2 SV=1 |
| Q8K157 | 1.18e-75 | 655 | 986 | 22 | 341 | Galactose mutarotase OS=Mus musculus OX=10090 GN=Galm PE=1 SV=1 |
| Q9GKX6 | 2.20e-74 | 655 | 986 | 22 | 341 | Galactose mutarotase OS=Sus scrofa OX=9823 GN=GALM PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
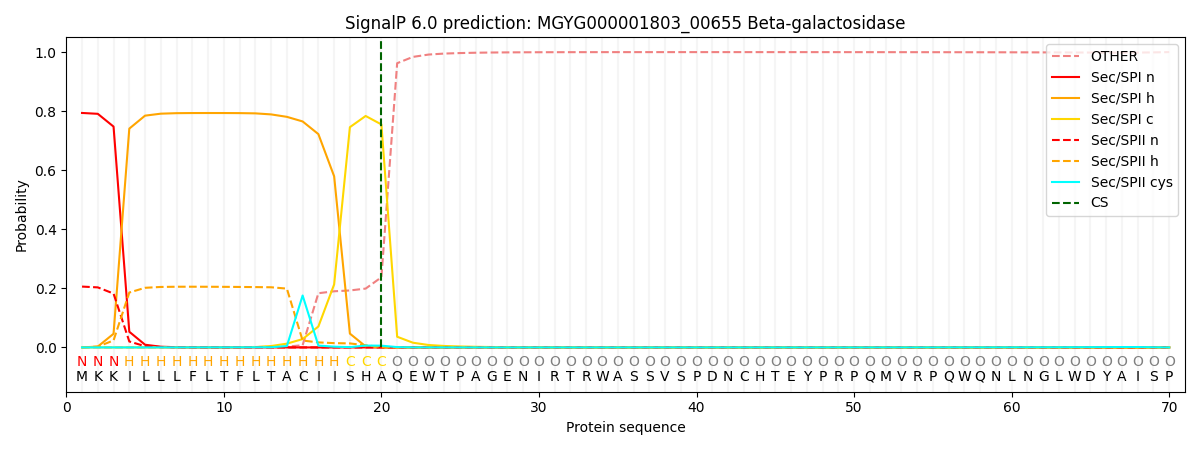
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000716 | 0.784616 | 0.213843 | 0.000302 | 0.000268 | 0.000246 |
