You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001804_01857
You are here: Home > Sequence: MGYG000001804_01857
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; CAG-831; | |||||||||||
| CAZyme ID | MGYG000001804_01857 | |||||||||||
| CAZy Family | GH53 | |||||||||||
| CAZyme Description | Arabinogalactan endo-beta-1,4-galactanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1427; End: 2533 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH53 | 45 | 366 | 3.6e-98 | 0.9941520467836257 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07745 | Glyco_hydro_53 | 4.67e-98 | 44 | 366 | 1 | 333 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| COG3867 | GanB | 2.74e-85 | 43 | 367 | 39 | 392 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
| smart00922 | MR_MLE | 0.007 | 71 | 121 | 7 | 53 | Mandelate racemase / muconate lactonizing enzyme, C-terminal domain. Mandelate racemase (MR) and muconate lactonizing enzyme (MLE) are two bacterial enzymes involved in aromatic acid catabolism. They catalyze mechanistically distinct reactions yet they are related at the level of their primary, quaternary (homooctamer) and tertiary structures.. This entry represents the C-terminal region of these proteins. |
| pfam13378 | MR_MLE_C | 0.008 | 77 | 168 | 1 | 88 | Enolase C-terminal domain-like. This domain appears at the C-terminus of many of the proteins that carry the MR_MLE_N pfam02746 domain. EC:4.2.1.40. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCD19010.1 | 3.85e-125 | 39 | 367 | 35 | 344 |
| QIK60890.1 | 3.98e-125 | 12 | 368 | 8 | 346 |
| QIK55473.1 | 1.79e-123 | 43 | 367 | 40 | 345 |
| APU98489.1 | 1.14e-120 | 43 | 367 | 35 | 340 |
| QMV66380.1 | 1.31e-119 | 43 | 367 | 35 | 340 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GPA_A | 6.78e-94 | 40 | 367 | 4 | 314 | Beta-1,4-galactanasefrom Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482],6GPA_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482] |
| 6GP5_A | 2.28e-93 | 40 | 367 | 40 | 350 | Beta-1,4-galactanasefrom Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],6GP5_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 1HJS_A | 5.27e-38 | 45 | 367 | 5 | 330 | Structureof two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_B Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_C Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_D Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_A Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_B Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_C Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_D Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus] |
| 7OSK_A | 8.65e-38 | 43 | 300 | 50 | 301 | ChainA, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230],7OSK_B Chain B, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230] |
| 1HJQ_A | 3.88e-37 | 45 | 366 | 5 | 329 | Structureof two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48843 | 3.10e-51 | 40 | 334 | 3 | 315 | Uncharacterized protein in bgaB 5'region (Fragment) OS=Niallia circulans OX=1397 PE=3 SV=1 |
| P83692 | 2.88e-37 | 45 | 367 | 5 | 330 | Arabinogalactan endo-beta-1,4-galactanase OS=Thermothelomyces thermophilus OX=78579 PE=1 SV=1 |
| P83691 | 2.12e-36 | 45 | 366 | 5 | 329 | Arabinogalactan endo-beta-1,4-galactanase OS=Humicola insolens OX=34413 PE=1 SV=1 |
| Q0CTQ7 | 8.21e-36 | 45 | 336 | 19 | 317 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=galA PE=3 SV=1 |
| Q65CX5 | 4.47e-35 | 43 | 256 | 49 | 265 | Endo-beta-1,4-galactanase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=ganB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
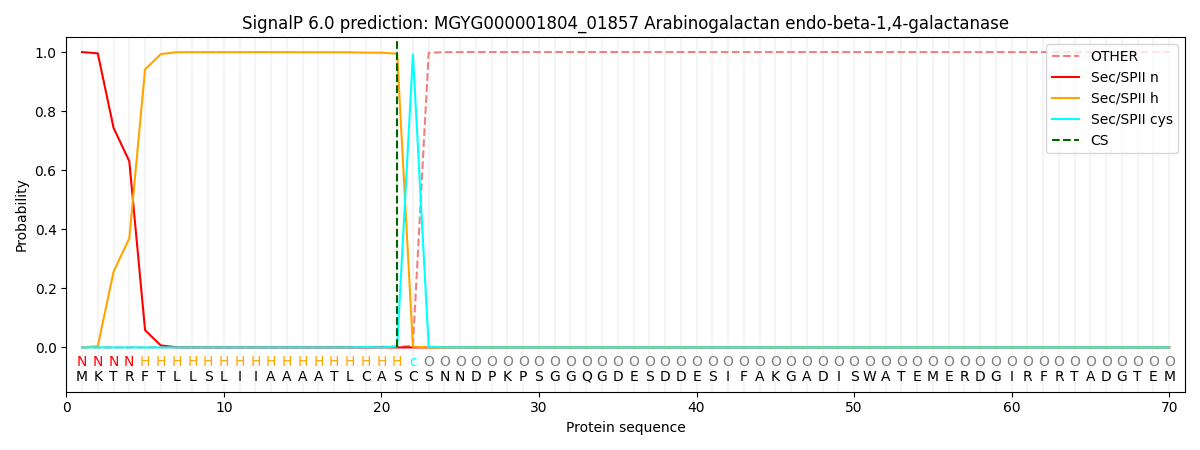
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000064 | 0.000000 | 0.000000 | 0.000000 |
