You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001818_00410
You are here: Home > Sequence: MGYG000001818_00410
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; QAKW01; HGM12957; | |||||||||||
| CAZyme ID | MGYG000001818_00410 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6; End: 5144 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 613 | 1350 | 4.9e-72 | 0.8361650485436893 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 4.99e-17 | 602 | 1116 | 338 | 829 | alpha-L-rhamnosidase. |
| NF033190 | inl_like_NEAT_1 | 9.97e-13 | 1552 | 1704 | 538 | 689 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 1.12e-09 | 1592 | 1632 | 1 | 41 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 1.08e-08 | 1530 | 1703 | 582 | 749 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 1.83e-08 | 1656 | 1698 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHF25191.1 | 4.05e-204 | 610 | 1388 | 252 | 1148 |
| QGH71057.1 | 2.57e-196 | 609 | 1388 | 237 | 1127 |
| QGH70002.1 | 6.35e-98 | 619 | 1387 | 280 | 1110 |
| SDG73971.1 | 6.58e-94 | 610 | 1379 | 172 | 965 |
| QNP57182.1 | 1.25e-93 | 608 | 1356 | 173 | 937 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 6.96e-07 | 609 | 1342 | 402 | 1092 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 7.33e-21 | 1528 | 1703 | 1680 | 1852 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 1.11e-19 | 1530 | 1703 | 908 | 1078 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P19424 | 1.03e-15 | 1530 | 1702 | 41 | 210 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| Q06853 | 2.76e-14 | 1549 | 1702 | 481 | 634 | Cell surface glycoprotein 2 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=Cthe_3079 PE=1 SV=1 |
| Q9ZES5 | 4.89e-13 | 1528 | 1709 | 32 | 208 | S-layer protein OS=Bacillus thuringiensis subsp. finitimus OX=29337 GN=ctc PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
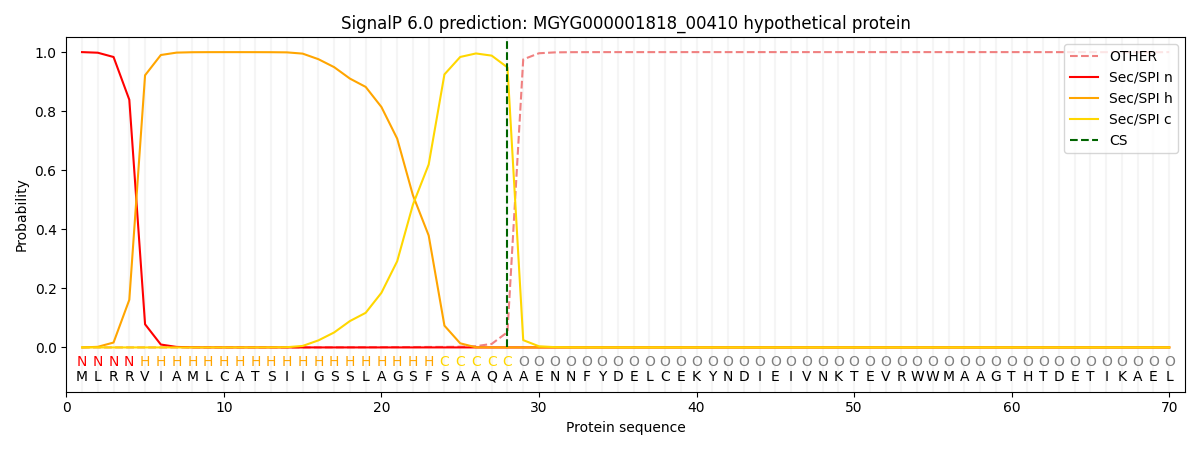
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000946 | 0.997834 | 0.000249 | 0.000487 | 0.000265 | 0.000206 |
