You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001851_00179
You are here: Home > Sequence: MGYG000001851_00179
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
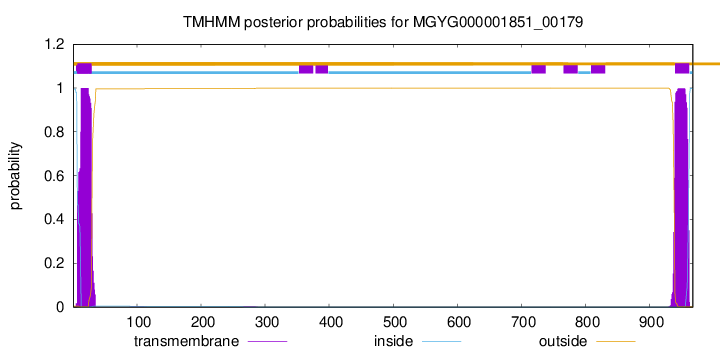
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-552; UMGS1880; | |||||||||||
| CAZyme ID | MGYG000001851_00179 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5464; End: 8370 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 668 | 859 | 2.5e-42 | 0.8935185185185185 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01915 | Glyco_hydro_3_C | 2.66e-26 | 82 | 337 | 2 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| COG1472 | BglX | 9.81e-23 | 654 | 871 | 46 | 268 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 2.29e-14 | 648 | 854 | 42 | 254 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK15098 | PRK15098 | 9.93e-11 | 76 | 464 | 390 | 723 | beta-glucosidase BglX. |
| pfam14310 | Fn3-like | 6.22e-10 | 423 | 505 | 1 | 71 | Fibronectin type III-like domain. This domain has a fibronectin type III-like structure. It is often found in association with pfam00933 and pfam01915. Its function is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOS39530.1 | 1.17e-311 | 1 | 967 | 1 | 964 |
| AIQ51426.1 | 1.08e-310 | 13 | 967 | 6 | 967 |
| AIQ45785.1 | 6.13e-310 | 13 | 967 | 6 | 967 |
| VEU79849.1 | 3.84e-286 | 1 | 967 | 1 | 988 |
| VEG24749.1 | 2.25e-216 | 6 | 965 | 8 | 1000 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 9.05e-56 | 79 | 896 | 46 | 784 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X40_A | 3.68e-30 | 659 | 904 | 35 | 294 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 2X42_A | 3.47e-29 | 659 | 904 | 35 | 294 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 4I3G_A | 1.21e-23 | 673 | 928 | 90 | 330 | CrystalStructure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae],4I3G_B Crystal Structure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae] |
| 5WAB_A | 9.64e-23 | 662 | 858 | 24 | 236 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15885 | 2.37e-57 | 76 | 854 | 16 | 696 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| P16084 | 7.34e-51 | 79 | 854 | 38 | 769 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P27034 | 1.81e-24 | 673 | 882 | 32 | 246 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| Q2U8Y5 | 1.17e-22 | 647 | 872 | 8 | 239 | Probable beta-glucosidase I OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglI PE=3 SV=1 |
| B8NDE2 | 1.17e-22 | 647 | 872 | 8 | 239 | Probable beta-glucosidase I OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglI PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
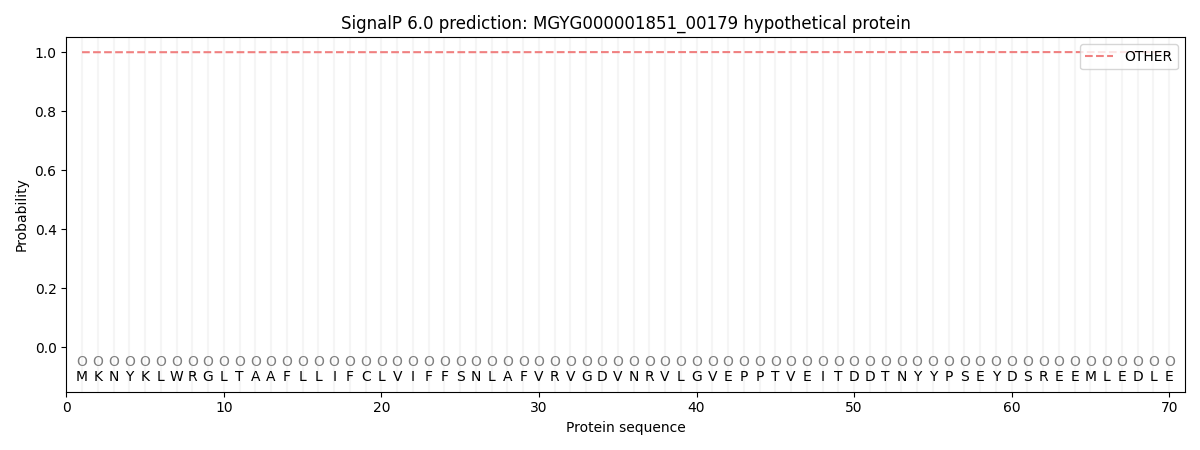
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999650 | 0.000331 | 0.000021 | 0.000001 | 0.000001 | 0.000019 |