You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001908_02142
You are here: Home > Sequence: MGYG000001908_02142
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_D sp900543145 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_D; Ruminococcus_D sp900543145 | |||||||||||
| CAZyme ID | MGYG000001908_02142 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 32117; End: 33292 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 67 | 355 | 3.4e-44 | 0.980327868852459 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd21510 | agarase_cat | 0.008 | 163 | 293 | 58 | 187 | alpha-beta barrel catalytic domain of agarase, such as GH86-like endo-acting agarases identified in non-marine organisms. Typically, agarases (E.C. 3.2.1.81) are found in ocean-dwelling bacteria since agarose is a principle component of red algae cell wall polysaccharides. Agarose is a linear polymer of alternating D-galactose and 3,6-anhydro-L-galactopyranose. Endo-acting agarases, such as glycoside hydrolase 16 (GH16) and GH86 hydrolyze internal beta-1,4 linkages. GH86-like endo-acting agarase of this protein family has been identified in the human intestinal bacterium Bacteroides uniformis. This acquired metabolic pathway, as demonstrated by the prevalence of agar-specific genetic cluster called polysaccharide utilization loci (PULs), varies considerably between human populations, being much more prevalent in a Japanese sample than in North America, European, or Chinese samples. Agarase activity was also identified in the non-marine bacterium Cellvibrio sp. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16471.1 | 4.93e-124 | 21 | 390 | 1 | 365 |
| ADD61974.1 | 2.22e-110 | 42 | 388 | 59 | 407 |
| CCO03823.1 | 6.68e-110 | 21 | 388 | 3 | 398 |
| AGS51602.1 | 3.27e-104 | 54 | 386 | 45 | 388 |
| ADU21807.1 | 1.42e-97 | 47 | 388 | 68 | 409 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZIZ_A | 2.62e-08 | 50 | 348 | 11 | 300 | ChainA, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
| 1RH9_A | 2.45e-06 | 55 | 214 | 7 | 179 | ChainA, endo-beta-mannanase [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2B3C0 | 2.50e-08 | 21 | 348 | 1 | 314 | Mannan endo-1,4-beta-mannosidase A OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=Pa_6_490 PE=1 SV=1 |
| B8NVK8 | 3.48e-08 | 24 | 214 | 13 | 209 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manA PE=3 SV=2 |
| Q2TXJ2 | 4.63e-08 | 24 | 214 | 13 | 209 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
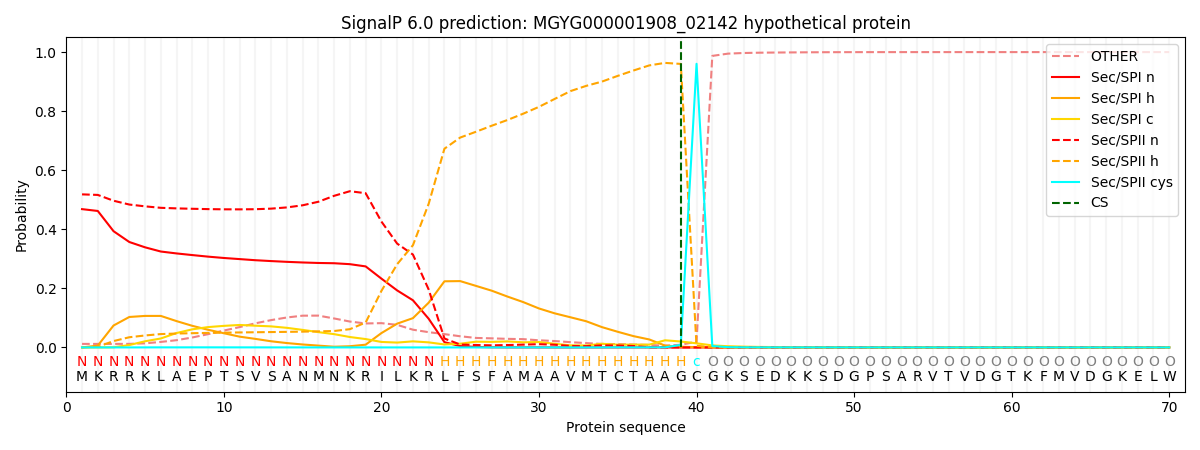
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003429 | 0.179215 | 0.816889 | 0.000300 | 0.000163 | 0.000022 |
