You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001920_00554
You are here: Home > Sequence: MGYG000001920_00554
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900546355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900546355 | |||||||||||
| CAZyme ID | MGYG000001920_00554 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34764; End: 35900 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 32 | 370 | 9.3e-106 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 5.33e-126 | 32 | 363 | 1 | 303 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 6.26e-109 | 75 | 368 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.76e-92 | 43 | 370 | 39 | 342 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAD80892.1 | 1.36e-208 | 1 | 371 | 1 | 372 |
| AVM57876.1 | 4.57e-191 | 1 | 370 | 1 | 370 |
| AVM52700.1 | 1.07e-189 | 1 | 372 | 1 | 372 |
| ALJ61519.1 | 3.89e-189 | 1 | 370 | 1 | 369 |
| QRQ48270.1 | 2.09e-188 | 22 | 370 | 25 | 369 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1UQY_A | 2.40e-99 | 26 | 362 | 21 | 359 | XylanaseXyn10B mutant (E262S) from Cellvibrio mixtus in complex with xylopentaose [Cellvibrio mixtus],1UQZ_A Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with 4-O-methyl glucuronic acid [Cellvibrio mixtus],1UR1_A Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha-1,3 linked to xylobiose [Cellvibrio mixtus],1UR2_A Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha 1,3 linked to xylotriose [Cellvibrio mixtus] |
| 2CNC_A | 1.40e-97 | 26 | 362 | 30 | 368 | Family10 xylanase [Cellvibrio mixtus] |
| 6FHE_A | 2.54e-97 | 32 | 363 | 13 | 333 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 4PMV_A | 7.25e-96 | 29 | 369 | 1 | 351 | Crystalstructure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P43212) [Xanthomonas citri pv. citri str. 306],4PMV_B Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P43212) [Xanthomonas citri pv. citri str. 306] |
| 4PMU_A | 3.98e-95 | 32 | 369 | 3 | 350 | Crystalstructure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) [Xanthomonas citri pv. citri str. 306],4PMU_B Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) [Xanthomonas citri pv. citri str. 306],4PMU_C Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) [Xanthomonas citri pv. citri str. 306],4PMU_D Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) [Xanthomonas citri pv. citri str. 306],4PMU_E Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) [Xanthomonas citri pv. citri str. 306],4PMU_F Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49942 | 2.64e-163 | 1 | 373 | 1 | 373 | Endo-1,4-beta-xylanase A OS=Bacteroides ovatus OX=28116 GN=xylI PE=2 SV=1 |
| P48789 | 6.11e-121 | 32 | 369 | 26 | 365 | Endo-1,4-beta-xylanase A OS=Prevotella ruminicola OX=839 GN=xynA PE=3 SV=1 |
| P45703 | 1.52e-79 | 32 | 363 | 8 | 322 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 GN=xynA PE=1 SV=1 |
| Q12603 | 5.20e-74 | 32 | 369 | 35 | 350 | Beta-1,4-xylanase OS=Dictyoglomus thermophilum OX=14 GN=xynA PE=3 SV=1 |
| O69231 | 5.73e-74 | 37 | 362 | 13 | 322 | Endo-1,4-beta-xylanase B OS=Paenibacillus barcinonensis OX=198119 GN=xynB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
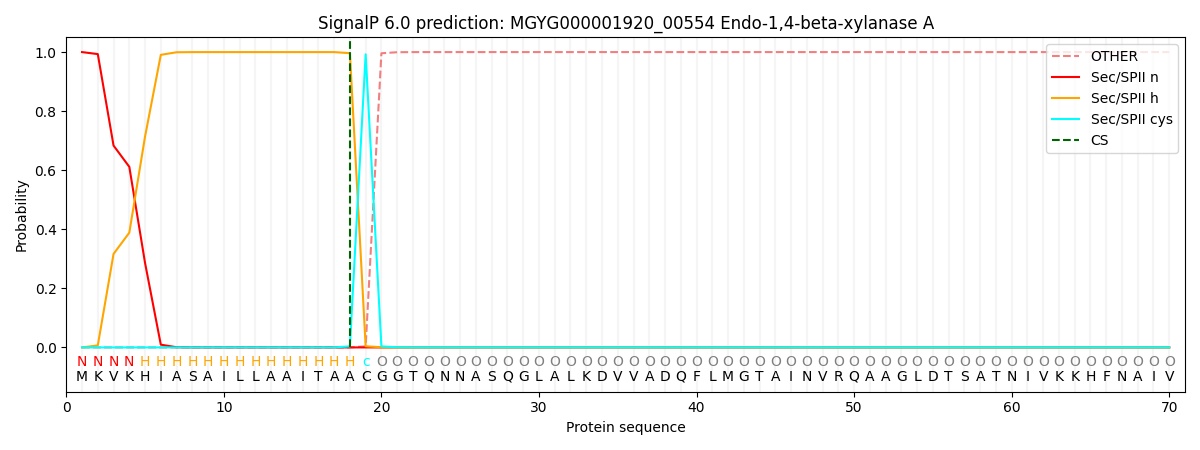
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000052 | 0.000000 | 0.000000 | 0.000000 |
