You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001920_01368
You are here: Home > Sequence: MGYG000001920_01368
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900546355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900546355 | |||||||||||
| CAZyme ID | MGYG000001920_01368 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8765; End: 9775 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 63 | 285 | 9.1e-51 | 0.5907692307692308 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.35e-55 | 30 | 291 | 73 | 373 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN03010 | PLN03010 | 1.60e-23 | 37 | 295 | 45 | 272 | polygalacturonase |
| PLN02218 | PLN02218 | 1.90e-21 | 34 | 250 | 63 | 271 | polygalacturonase ADPG |
| PLN02188 | PLN02188 | 2.54e-20 | 35 | 294 | 33 | 301 | polygalacturonase/glycoside hydrolase family protein |
| PLN03003 | PLN03003 | 7.36e-18 | 40 | 250 | 25 | 217 | Probable polygalacturonase At3g15720 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY36938.1 | 2.50e-186 | 14 | 291 | 6 | 283 |
| QDO68557.1 | 3.79e-161 | 13 | 291 | 1 | 281 |
| ALJ60362.1 | 2.18e-160 | 13 | 291 | 1 | 281 |
| QUT88631.1 | 4.38e-160 | 13 | 291 | 1 | 281 |
| QJR78587.1 | 6.95e-157 | 29 | 288 | 19 | 278 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 3.00e-25 | 32 | 282 | 33 | 312 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3JUR_A | 1.36e-18 | 32 | 253 | 20 | 270 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 2UVE_A | 5.78e-14 | 32 | 246 | 153 | 405 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
| 4C2L_A | 1.85e-10 | 46 | 287 | 23 | 248 | Crystalstructure of endo-xylogalacturonan hydrolase from Aspergillus tubingensis [Aspergillus tubingensis] |
| 2PYG_A | 4.40e-08 | 39 | 78 | 3 | 42 | Azotobactervinelandii Mannuronan C-5 epimerase AlgE4 A-module [Azotobacter vinelandii],2PYG_B Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module [Azotobacter vinelandii],2PYH_A Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module complexed with mannuronan trisaccharide [Azotobacter vinelandii],2PYH_B Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module complexed with mannuronan trisaccharide [Azotobacter vinelandii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7PZL3 | 1.02e-23 | 40 | 287 | 64 | 319 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
| Q8RY29 | 3.33e-19 | 40 | 250 | 69 | 271 | Polygalacturonase ADPG2 OS=Arabidopsis thaliana OX=3702 GN=ADPG2 PE=2 SV=2 |
| Q7M1E7 | 5.50e-17 | 32 | 295 | 52 | 298 | Polygalacturonase OS=Chamaecyparis obtusa OX=13415 PE=1 SV=1 |
| Q9LW07 | 2.08e-16 | 35 | 250 | 20 | 217 | Probable polygalacturonase At3g15720 OS=Arabidopsis thaliana OX=3702 GN=At3g15720 PE=3 SV=1 |
| P35339 | 1.89e-15 | 39 | 248 | 41 | 238 | Exopolygalacturonase OS=Zea mays OX=4577 GN=PG2C PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
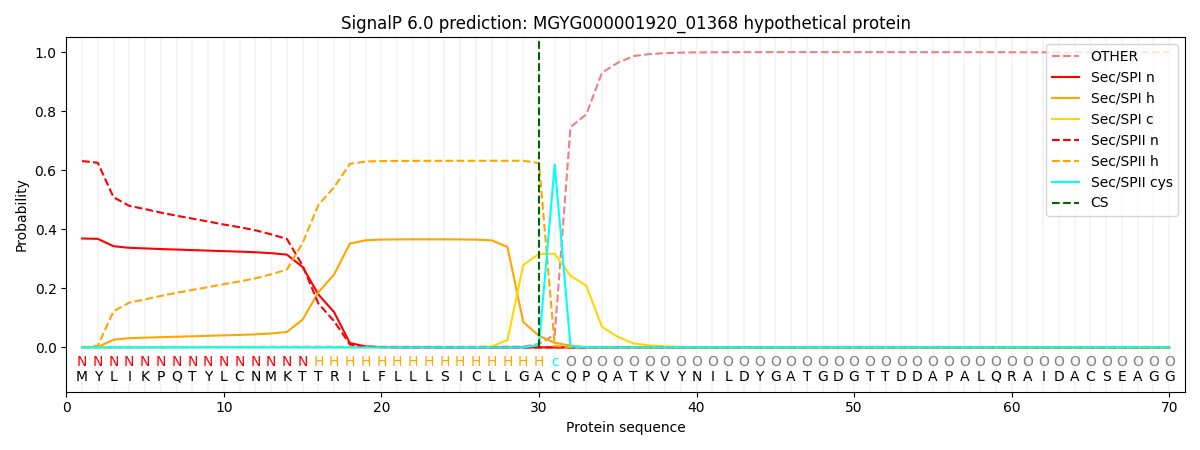
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000462 | 0.358739 | 0.640322 | 0.000157 | 0.000184 | 0.000131 |
