You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001926_00700
You are here: Home > Sequence: MGYG000001926_00700
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
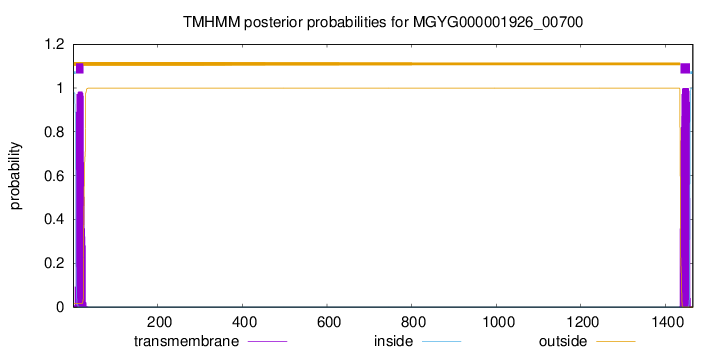
TMHMM annotations
Basic Information help
| Species | CAG-776 sp900549285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-776; CAG-776 sp900549285 | |||||||||||
| CAZyme ID | MGYG000001926_00700 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43070; End: 47467 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 74 | 209 | 2.4e-24 | 0.9104477611940298 |
| CBM32 | 886 | 994 | 2.2e-18 | 0.8145161290322581 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 2.04e-29 | 48 | 210 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 2.43e-20 | 50 | 209 | 5 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam00754 | F5_F8_type_C | 1.69e-11 | 888 | 997 | 7 | 119 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam13285 | DUF4073 | 0.002 | 808 | 849 | 97 | 141 | Domain of unknown function (DUF4073). This family is frequently found at the C-terminus of bacterial proteins carrying the family, Metallophos pfam00149. |
| pfam17002 | DUF5089 | 0.005 | 1221 | 1302 | 101 | 193 | Domain of unknown function (DUF5089). This is a family of microsporidial-specific proteins of unknown function. There is distant homology to synaptosomal-associated 25 family proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCL58565.1 | 0.0 | 48 | 1156 | 79 | 1191 |
| QGM22805.1 | 2.45e-174 | 48 | 1061 | 138 | 1139 |
| SQG16429.1 | 3.65e-174 | 48 | 1014 | 141 | 1099 |
| VED84782.1 | 3.65e-174 | 48 | 1014 | 141 | 1099 |
| QZA21247.1 | 1.28e-173 | 48 | 1061 | 138 | 1139 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VMI_A | 3.49e-08 | 50 | 209 | 9 | 150 | Thestructure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMH_A | 3.49e-08 | 50 | 209 | 9 | 150 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMG_A | 3.96e-08 | 50 | 209 | 15 | 156 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
| 2EIC_A | 1.20e-07 | 879 | 990 | 11 | 128 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 2EIB_A | 1.20e-07 | 879 | 990 | 11 | 128 | ChainA, Galactose oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| I1S2N3 | 2.99e-07 | 842 | 990 | 17 | 169 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
| P0CS93 | 6.78e-07 | 879 | 990 | 52 | 169 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
| Q02834 | 7.70e-06 | 897 | 990 | 528 | 626 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
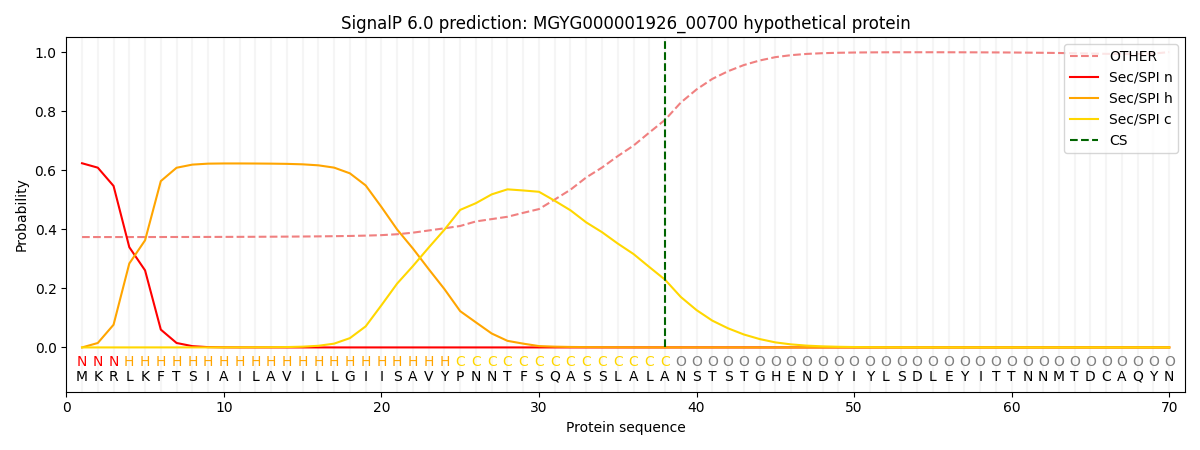
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.397464 | 0.594688 | 0.003625 | 0.001812 | 0.000697 | 0.001701 |