You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001940_01387
You are here: Home > Sequence: MGYG000001940_01387
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp002438605 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp002438605 | |||||||||||
| CAZyme ID | MGYG000001940_01387 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase Y | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 23273; End: 26461 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 186 | 531 | 1.6e-81 | 0.9867986798679867 |
| CE1 | 827 | 1057 | 1.6e-30 | 0.9427312775330396 |
| CBM22 | 557 | 686 | 8.3e-30 | 0.9694656488549618 |
| CBM22 | 29 | 154 | 1e-15 | 0.9618320610687023 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 7.68e-93 | 186 | 530 | 1 | 308 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 5.84e-81 | 228 | 530 | 2 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.11e-50 | 196 | 535 | 36 | 342 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam02018 | CBM_4_9 | 5.07e-17 | 554 | 690 | 1 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| COG2382 | Fes | 9.06e-12 | 803 | 1060 | 55 | 296 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUG56905.1 | 0.0 | 1 | 1061 | 7 | 1086 |
| ABN52146.1 | 0.0 | 1 | 1061 | 7 | 1076 |
| ADU74368.1 | 0.0 | 1 | 1061 | 7 | 1076 |
| ALX08313.1 | 0.0 | 1 | 1061 | 7 | 1076 |
| ANV76061.1 | 0.0 | 1 | 1061 | 7 | 1076 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W5F_A | 1.44e-210 | 28 | 544 | 24 | 538 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2WYS_A | 4.07e-207 | 28 | 544 | 24 | 538 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 1GKL_A | 2.99e-130 | 802 | 1061 | 27 | 288 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1GKL_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1WB4_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB4_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB5_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB5_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB6_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1WB6_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 6FJ4_A | 1.49e-123 | 802 | 1061 | 13 | 274 | ChainA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus] |
| 1GKK_A | 2.44e-123 | 802 | 1061 | 27 | 288 | ChainA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1GKK_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],3ZI7_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus YS],3ZI7_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus YS],4BAG_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],4BAG_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],4H35_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],4H35_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],5FXM_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],6Y8G_AAA Chain AAA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],6Y8G_BBB Chain BBB, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P51584 | 0.0 | 1 | 1061 | 7 | 1076 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| P29126 | 3.49e-79 | 179 | 533 | 623 | 952 | Bifunctional endo-1,4-beta-xylanase XylA OS=Ruminococcus flavefaciens OX=1265 GN=xynA PE=3 SV=1 |
| Q60042 | 2.67e-55 | 29 | 545 | 201 | 706 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| Q60037 | 3.45e-53 | 29 | 545 | 206 | 710 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| P26223 | 2.99e-49 | 186 | 509 | 3 | 309 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
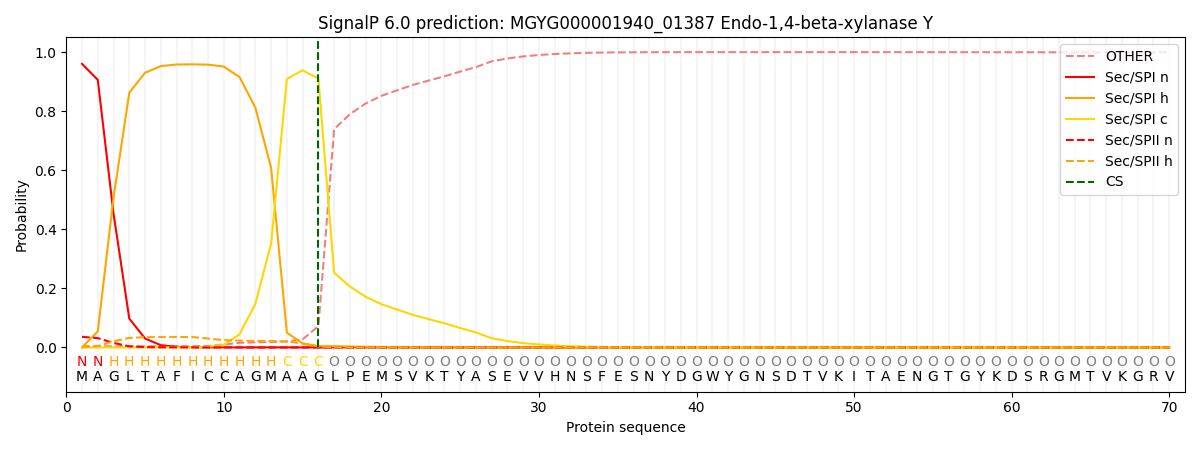
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005785 | 0.951284 | 0.041905 | 0.000391 | 0.000297 | 0.000296 |
