You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002020_01096
You are here: Home > Sequence: MGYG000002020_01096
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Collinsella sp900544225 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; Collinsella sp900544225 | |||||||||||
| CAZyme ID | MGYG000002020_01096 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 82120; End: 84198 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 38 | 525 | 3.8e-39 | 0.7425925925925926 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 2.46e-24 | 35 | 525 | 2 | 396 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 1.01e-22 | 96 | 259 | 1 | 152 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1287 | Stt3 | 0.003 | 79 | 439 | 76 | 436 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
| COG5305 | COG5305 | 0.005 | 96 | 257 | 96 | 258 | Uncharacterized membrane protein [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWZ09319.1 | 1.90e-89 | 40 | 409 | 36 | 380 |
| QUB37647.1 | 4.90e-88 | 46 | 689 | 210 | 792 |
| QHU89858.1 | 9.50e-88 | 40 | 692 | 273 | 904 |
| QHU90800.1 | 2.99e-80 | 40 | 692 | 273 | 901 |
| QHU92512.1 | 4.14e-80 | 40 | 692 | 271 | 901 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34575 | 2.27e-64 | 41 | 675 | 10 | 660 | Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
| P37483 | 8.74e-54 | 42 | 675 | 9 | 647 | Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
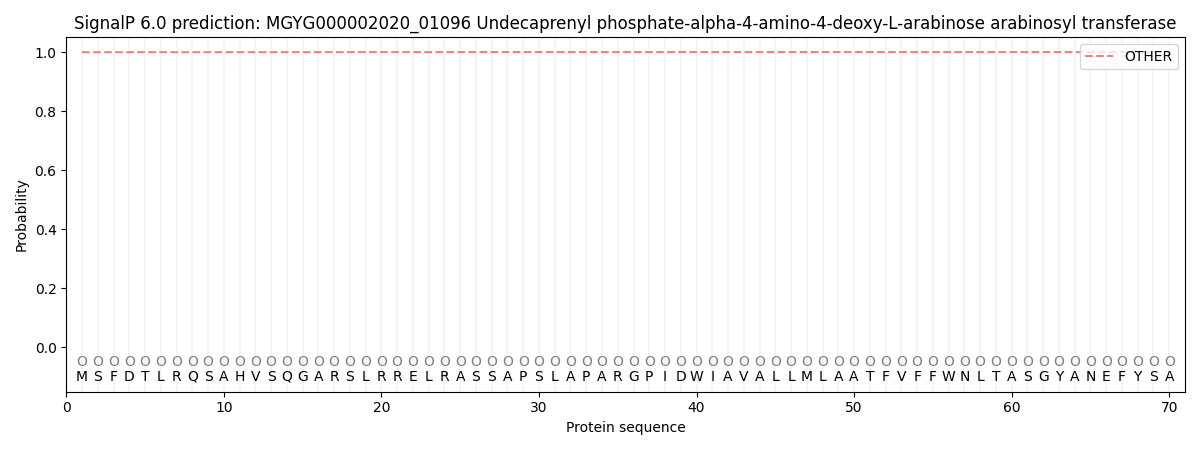
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999829 | 0.000171 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
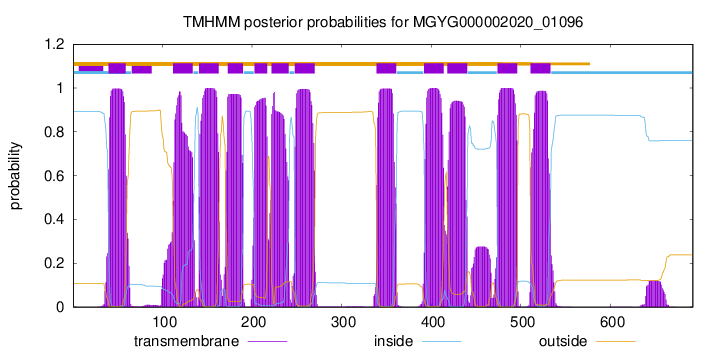
| start | end |
|---|---|
| 40 | 59 |
| 112 | 134 |
| 141 | 163 |
| 173 | 190 |
| 203 | 217 |
| 222 | 241 |
| 248 | 270 |
| 339 | 361 |
| 392 | 414 |
| 418 | 440 |
| 474 | 496 |
| 511 | 533 |
