You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002051_00864
You are here: Home > Sequence: MGYG000002051_00864
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900546345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900546345 | |||||||||||
| CAZyme ID | MGYG000002051_00864 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20511; End: 22196 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 282 | 556 | 9.3e-48 | 0.7260726072607261 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.43e-38 | 307 | 556 | 96 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 5.45e-36 | 304 | 554 | 51 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.09e-19 | 304 | 561 | 117 | 344 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam02018 | CBM_4_9 | 0.007 | 171 | 241 | 53 | 128 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAB01855.1 | 9.33e-298 | 3 | 559 | 5 | 558 |
| BCS85403.1 | 7.42e-196 | 2 | 561 | 4 | 561 |
| QUT92912.1 | 1.09e-150 | 8 | 561 | 13 | 730 |
| QUT32420.1 | 6.00e-145 | 12 | 561 | 8 | 746 |
| QRN02128.1 | 6.00e-145 | 12 | 561 | 8 | 746 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGQ_A | 1.28e-77 | 135 | 304 | 1 | 168 | PbXyn10CCBM APO [Prevotella bryantii B14] |
| 1W32_A | 5.99e-19 | 328 | 561 | 118 | 348 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W32_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W3H_A | 6.80e-19 | 328 | 561 | 129 | 359 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W3H_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1I1W_A | 1.47e-18 | 308 | 559 | 101 | 302 | 0.89AUltra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus [Thermoascus aurantiacus],1I1X_A 1.11 A ATOMIC RESOLUTION STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS [Thermoascus aurantiacus] |
| 1CLX_A | 2.60e-18 | 328 | 561 | 117 | 347 | CatalyticCore Of Xylanase A [Cellvibrio japonicus],1CLX_B Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_C Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_D Catalytic Core Of Xylanase A [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q96VB6 | 7.98e-21 | 301 | 558 | 119 | 323 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
| Q4JHP5 | 1.01e-18 | 301 | 558 | 122 | 326 | Probable endo-1,4-beta-xylanase C OS=Aspergillus terreus OX=33178 GN=xlnC PE=2 SV=1 |
| Q0CBM8 | 1.37e-18 | 301 | 558 | 122 | 326 | Probable endo-1,4-beta-xylanase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=xlnC PE=2 SV=2 |
| W0HFK8 | 6.48e-18 | 308 | 558 | 131 | 331 | Endo-1,4-beta-xylanase 1 OS=Rhizopus oryzae OX=64495 GN=xyn1 PE=1 SV=1 |
| Q6PRW6 | 2.05e-17 | 308 | 551 | 131 | 324 | Endo-1,4-beta-xylanase OS=Penicillium chrysogenum OX=5076 GN=Xyn PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
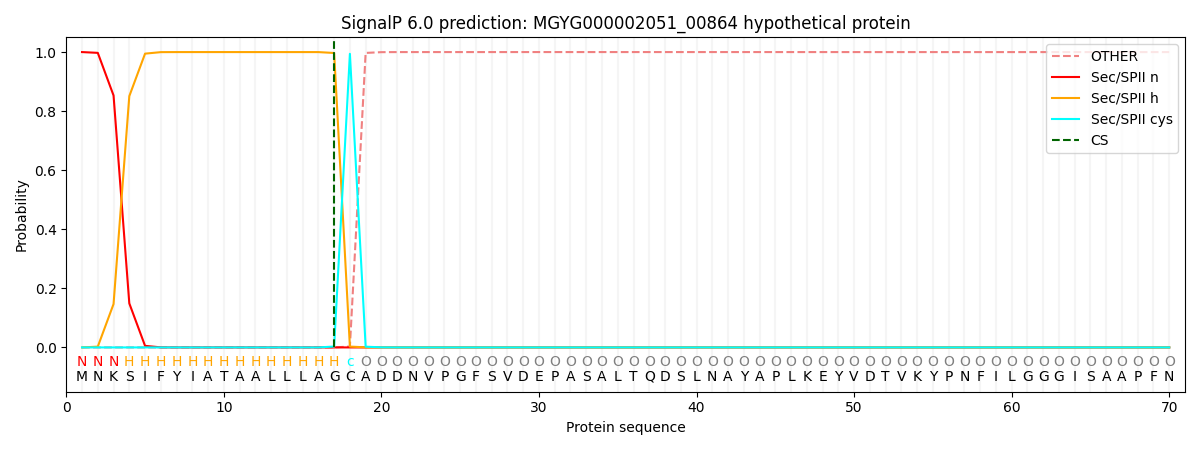
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000061 | 0.000000 | 0.000000 | 0.000000 |
