You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002132_01713
You are here: Home > Sequence: MGYG000002132_01713
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Monoglobus sp900542675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Monoglobaceae; Monoglobus; Monoglobus sp900542675 | |||||||||||
| CAZyme ID | MGYG000002132_01713 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 305; End: 3634 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 689 | 911 | 1.8e-45 | 0.9103139013452914 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1928 | PMT1 | 2.57e-17 | 920 | 1105 | 495 | 696 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG1928 | PMT1 | 1.92e-15 | 675 | 910 | 49 | 264 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG5542 | COG5542 | 1.07e-14 | 107 | 404 | 92 | 369 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| pfam16192 | PMT_4TMC | 1.18e-14 | 930 | 1100 | 7 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| pfam13231 | PMT_2 | 2.02e-07 | 116 | 264 | 4 | 155 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO18475.1 | 0.0 | 1 | 1107 | 1 | 1160 |
| AIQ66842.1 | 1.18e-184 | 56 | 1104 | 11 | 1031 |
| AIQ15817.1 | 3.53e-183 | 56 | 1104 | 11 | 1031 |
| AIQ44967.1 | 5.91e-183 | 56 | 1104 | 6 | 1025 |
| CQR52353.1 | 3.33e-182 | 56 | 1104 | 255 | 1275 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WN04 | 3.22e-19 | 717 | 1102 | 102 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 3.22e-19 | 717 | 1102 | 102 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| L8F4Z2 | 6.66e-17 | 694 | 1102 | 67 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| Q8NRZ6 | 8.45e-12 | 717 | 1102 | 99 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000018 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
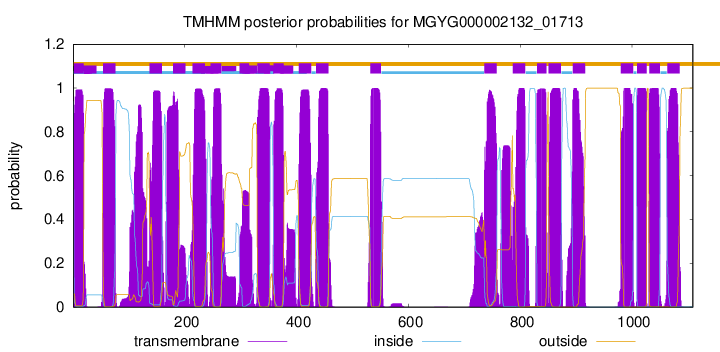
| start | end |
|---|---|
| 2 | 19 |
| 54 | 76 |
| 137 | 159 |
| 179 | 201 |
| 214 | 236 |
| 246 | 265 |
| 298 | 315 |
| 330 | 352 |
| 359 | 377 |
| 404 | 426 |
| 435 | 457 |
| 532 | 551 |
| 736 | 758 |
| 787 | 809 |
| 830 | 847 |
| 851 | 873 |
| 894 | 916 |
| 980 | 1002 |
| 1009 | 1026 |
| 1031 | 1050 |
| 1063 | 1085 |
