You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002164_00368
You are here: Home > Sequence: MGYG000002164_00368
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Methanosphaera stadtmanae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Archaea; Methanobacteriota; Methanobacteria; Methanobacteriales; Methanobacteriaceae; Methanosphaera; Methanosphaera stadtmanae | |||||||||||
| CAZyme ID | MGYG000002164_00368 | |||||||||||
| CAZy Family | GT66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 433673; End: 436249 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT66 | 22 | 701 | 1e-152 | 0.9393939393939394 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02516 | STT3 | 8.16e-53 | 27 | 550 | 4 | 478 | Oligosaccharyl transferase STT3 subunit. This family consists of the oligosaccharyl transferase STT3 subunit and related proteins. The STT3 subunit is part of the oligosaccharyl transferase (OTase) complex of proteins and is required for its activity. In eukaryotes, OTase transfers a lipid-linked core-oligosaccharide to selected asparagine residues in the ER. In the archaea STT3 occurs alone, rather than in an OTase complex, and is required for N-glycosylation of asparagines. |
| COG1287 | Stt3 | 3.86e-50 | 10 | 665 | 16 | 673 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABC56771.1 | 0.0 | 1 | 858 | 1 | 858 |
| AWX32536.1 | 0.0 | 1 | 838 | 1 | 839 |
| AEG18866.1 | 7.11e-207 | 1 | 832 | 1 | 821 |
| SCG86282.1 | 1.66e-206 | 1 | 832 | 1 | 815 |
| ADZ08786.1 | 1.21e-197 | 1 | 832 | 1 | 816 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FTJ_5 | 7.15e-09 | 507 | 611 | 503 | 601 | Cryo-EMStructure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome [Canis lupus familiaris] |
| 6FTG_5 | 7.20e-09 | 507 | 611 | 503 | 601 | Subtomogramaverage of OST-containing ribosome-translocon complexes from canine rough microsomal membranes [Canis lupus familiaris],6FTI_5 Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome [Canis lupus familiaris] |
| 6S7O_A | 7.20e-09 | 507 | 611 | 503 | 601 | Cryo-EMstructure of human oligosaccharyltransferase complex OST-A [Homo sapiens] |
| 6C26_A | 1.32e-06 | 507 | 576 | 494 | 562 | TheCryo-EM structure of a eukaryotic oligosaccharyl transferase complex [Saccharomyces cerevisiae S288C],6EZN_F Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex [Saccharomyces cerevisiae S288C],7OCI_F Chain F, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3 [Saccharomyces cerevisiae S288C] |
| 2LGZ_A | 2.06e-06 | 507 | 576 | 49 | 117 | Solutionstructure of STT3P [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2EMT4 | 5.85e-69 | 14 | 836 | 40 | 914 | Dolichyl-phosphooligosaccharide-protein glycotransferase OS=Methanococcus voltae OX=2188 GN=aglB PE=1 SV=1 |
| Q58920 | 4.67e-62 | 10 | 831 | 25 | 925 | Dolichyl-phosphooligosaccharide-protein glycotransferase OS=Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) OX=243232 GN=aglB PE=3 SV=1 |
| Q8U4D2 | 3.32e-10 | 53 | 549 | 50 | 533 | Dolichyl-phosphooligosaccharide-protein glycotransferase 1 OS=Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1) OX=186497 GN=aglB1 PE=1 SV=1 |
| Q2KJI2 | 3.94e-08 | 507 | 611 | 503 | 601 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A OS=Bos taurus OX=9913 GN=STT3A PE=2 SV=1 |
| Q5RCE2 | 3.94e-08 | 507 | 611 | 503 | 601 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A OS=Pongo abelii OX=9601 GN=STT3A PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.307909 | 0.679927 | 0.010939 | 0.000435 | 0.000347 | 0.000401 |
TMHMM Annotations download full data without filtering help
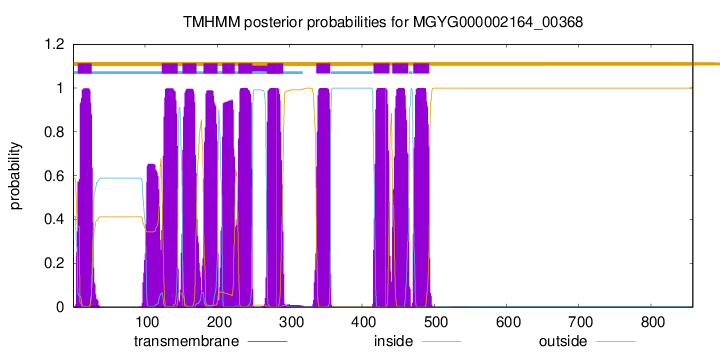
| start | end |
|---|---|
| 7 | 26 |
| 123 | 145 |
| 152 | 171 |
| 181 | 200 |
| 207 | 224 |
| 229 | 248 |
| 269 | 291 |
| 337 | 356 |
| 416 | 438 |
| 442 | 464 |
| 471 | 493 |
