You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002218_00311
You are here: Home > Sequence: MGYG000002218_00311
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
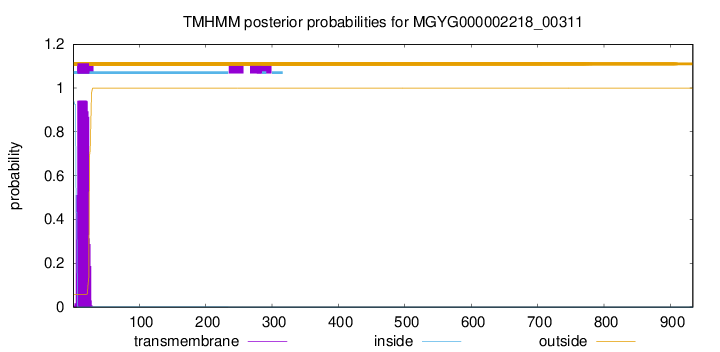
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp000436495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp000436495 | |||||||||||
| CAZyme ID | MGYG000002218_00311 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 114763; End: 117567 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 22 | 486 | 2.4e-72 | 0.4933510638297872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10150 | PRK10150 | 2.20e-27 | 18 | 343 | 7 | 296 | beta-D-glucuronidase; Provisional |
| COG3250 | LacZ | 1.59e-25 | 24 | 534 | 13 | 500 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 1.17e-19 | 26 | 469 | 44 | 470 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 1.91e-11 | 218 | 323 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02837 | Glyco_hydro_2_N | 2.47e-08 | 26 | 206 | 4 | 159 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT75482.1 | 0.0 | 26 | 933 | 26 | 932 |
| QRO25183.1 | 0.0 | 24 | 934 | 26 | 932 |
| AYB35766.1 | 0.0 | 22 | 933 | 48 | 968 |
| SCD20884.1 | 0.0 | 24 | 929 | 26 | 937 |
| SDS50045.1 | 0.0 | 13 | 934 | 13 | 946 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4JKM_A | 8.47e-20 | 24 | 469 | 16 | 441 | CrystalStructure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],4JKM_B Crystal Structure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],6CXS_A Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13],6CXS_B Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13] |
| 5C70_A | 7.71e-15 | 51 | 474 | 18 | 453 | Thestructure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae],5C70_B The structure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae] |
| 5C71_A | 1.85e-14 | 51 | 474 | 43 | 478 | Thestructure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_B The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_C The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_D The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae] |
| 7KGY_A | 6.99e-14 | 24 | 482 | 18 | 459 | ChainA, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_B Chain B, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_C Chain C, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_D Chain D, Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 6ED2_A | 3.11e-11 | 24 | 472 | 40 | 473 | ChainA, Glycosyl hydrolase family 2, TIM barrel domain protein [Faecalibacterium duncaniae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P06760 | 2.81e-18 | 26 | 480 | 41 | 492 | Beta-glucuronidase OS=Rattus norvegicus OX=10116 GN=Gusb PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
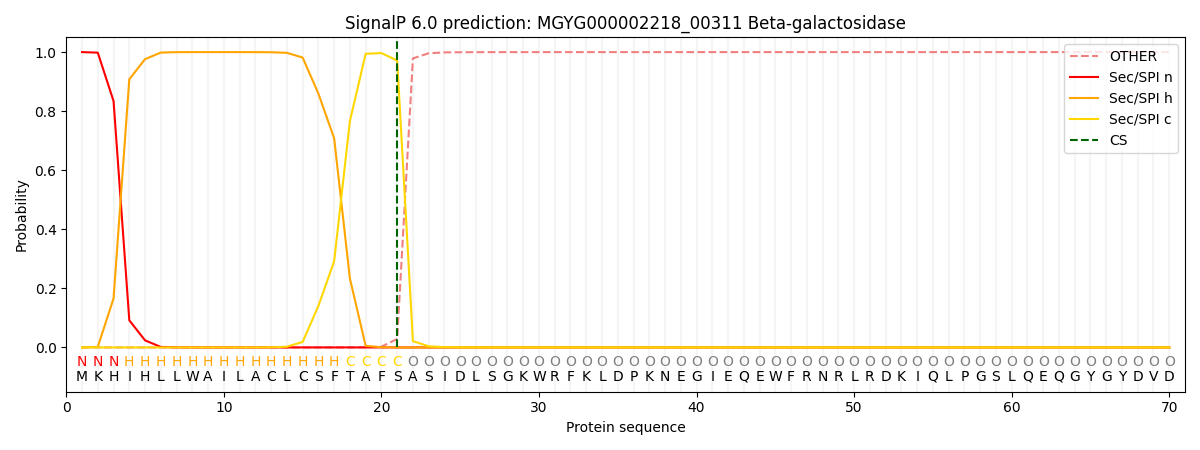
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000399 | 0.998739 | 0.000257 | 0.000199 | 0.000186 | 0.000191 |