You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002275_01296
You are here: Home > Sequence: MGYG000002275_01296
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella sp003477995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella sp003477995 | |||||||||||
| CAZyme ID | MGYG000002275_01296 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 84408; End: 85988 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 12 | 492 | 7.7e-57 | 0.9148148148148149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 6.12e-21 | 21 | 444 | 16 | 447 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 1.02e-04 | 21 | 338 | 15 | 328 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 0.005 | 68 | 227 | 1 | 155 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUU03965.1 | 2.88e-118 | 22 | 503 | 10 | 494 |
| QRP88934.1 | 3.23e-117 | 22 | 503 | 10 | 494 |
| QLK84536.1 | 7.21e-116 | 22 | 503 | 10 | 494 |
| QKH86812.1 | 1.02e-115 | 22 | 503 | 10 | 494 |
| QTO25703.1 | 1.44e-115 | 22 | 503 | 10 | 494 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4ZSZ0 | 1.36e-10 | 21 | 408 | 16 | 402 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=arnT PE=3 SV=1 |
| O67270 | 1.55e-09 | 30 | 347 | 24 | 328 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
| Q48HY9 | 8.22e-08 | 21 | 408 | 15 | 401 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas savastanoi pv. phaseolicola (strain 1448A / Race 6) OX=264730 GN=arnT PE=3 SV=1 |
| Q4KC80 | 1.93e-07 | 19 | 469 | 12 | 468 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=arnT1 PE=3 SV=1 |
| Q1CIH9 | 1.36e-06 | 19 | 327 | 14 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pestis bv. Antiqua (strain Nepal516) OX=377628 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
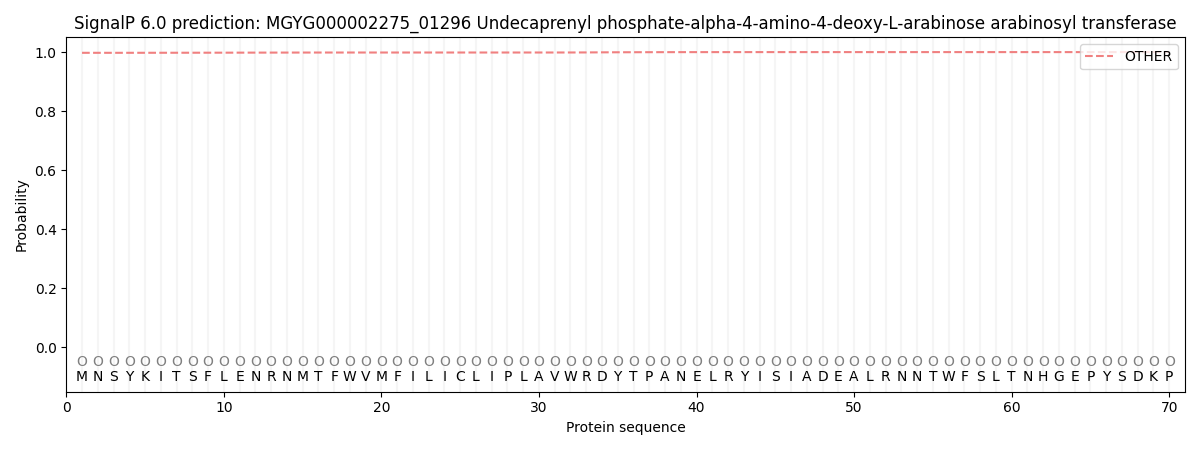
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.997687 | 0.001762 | 0.000074 | 0.000009 | 0.000007 | 0.000478 |
TMHMM Annotations download full data without filtering help
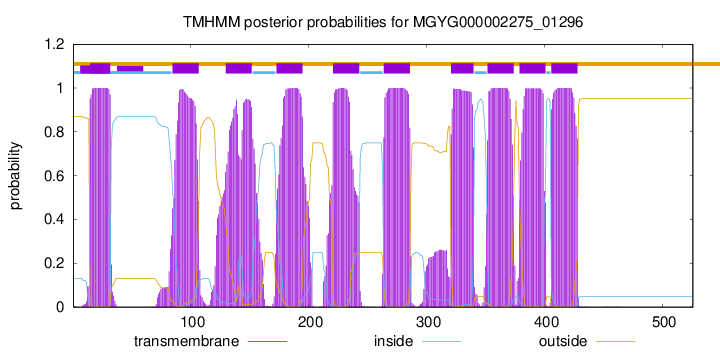
| start | end |
|---|---|
| 15 | 32 |
| 85 | 107 |
| 130 | 152 |
| 173 | 195 |
| 221 | 243 |
| 264 | 286 |
| 321 | 340 |
| 352 | 374 |
| 379 | 401 |
| 406 | 428 |
