You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002280_00571
You are here: Home > Sequence: MGYG000002280_00571
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA9502 sp003481825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UBA9502; UBA9502 sp003481825 | |||||||||||
| CAZyme ID | MGYG000002280_00571 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7876; End: 9045 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 121 | 361 | 4e-25 | 0.9580152671755725 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.62e-08 | 117 | 346 | 20 | 257 | Cellulase (glycosyl hydrolase family 5). |
| COG5263 | COG5263 | 1.01e-07 | 26 | 83 | 257 | 313 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| NF033838 | PspC_subgroup_1 | 1.74e-05 | 21 | 113 | 543 | 630 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
| NF033838 | PspC_subgroup_1 | 2.16e-05 | 23 | 83 | 625 | 683 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
| NF033840 | PspC_relate_1 | 2.84e-05 | 21 | 99 | 527 | 601 | PspC-related protein choline-binding protein 1. Members of this family share C-terminal homology to the choline-binding form of the pneumococcal surface antigen PspC, but not to its allelic LPXTG-anchored forms because they lack the choline-binding repeat region. Members of this family should not be confused with PspC itself, whose identity and function reflect regions N-terminal to the choline-binding region. See Iannelli, et al. (PMID: 11891047) for information about the different allelic forms of PspC. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIB28168.1 | 5.54e-144 | 91 | 388 | 2 | 299 |
| QCI58964.2 | 3.55e-138 | 91 | 382 | 28 | 319 |
| QMW91197.1 | 1.03e-129 | 93 | 382 | 28 | 317 |
| BBK76626.1 | 1.03e-129 | 93 | 382 | 28 | 317 |
| QUF84801.1 | 1.46e-129 | 93 | 382 | 28 | 317 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3HZ3_A | 2.90e-06 | 26 | 83 | 903 | 960 | Lactobacillusreuteri N-terminally truncated glucansucrase GTF180(D1025N)-sucrose complex [Limosilactobacillus reuteri] |
| 3KLK_A | 2.90e-06 | 26 | 83 | 903 | 960 | Crystalstructure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in triclinic apo- form [Limosilactobacillus reuteri],4AYG_A Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in orthorhombic apo-form [Limosilactobacillus reuteri],4AYG_B Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in orthorhombic apo-form [Limosilactobacillus reuteri] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
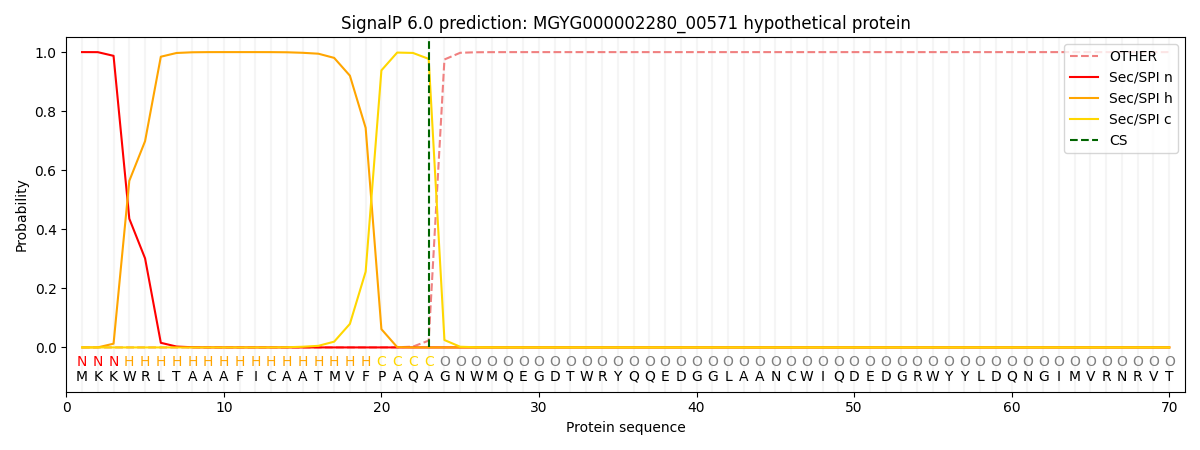
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000295 | 0.998884 | 0.000267 | 0.000193 | 0.000187 | 0.000167 |
