You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002281_02711
You are here: Home > Sequence: MGYG000002281_02711
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
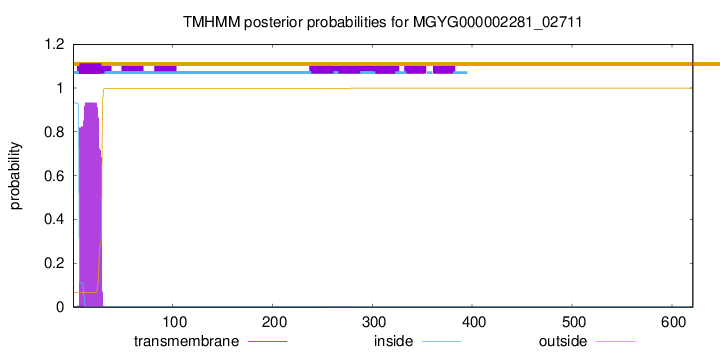
TMHMM annotations
Basic Information help
| Species | Bacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides faecis | |||||||||||
| CAZyme ID | MGYG000002281_02711 | |||||||||||
| CAZy Family | CBM62 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 98029; End: 99894 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM62 | 464 | 535 | 2.5e-24 | 0.5419847328244275 |
| CBM62 | 543 | 602 | 1.4e-18 | 0.45038167938931295 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT71327.1 | 0.0 | 1 | 621 | 1 | 621 |
| BCA50427.1 | 0.0 | 1 | 621 | 1 | 621 |
| QQA06265.1 | 0.0 | 1 | 621 | 1 | 621 |
| QIU96201.1 | 0.0 | 1 | 620 | 1 | 616 |
| QDH56165.1 | 0.0 | 1 | 620 | 1 | 619 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2YB7_A | 1.47e-08 | 468 | 538 | 70 | 146 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YB7_B Chain B, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YFU_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YFZ_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YG0_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5G56_A | 5.17e-07 | 468 | 542 | 771 | 851 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
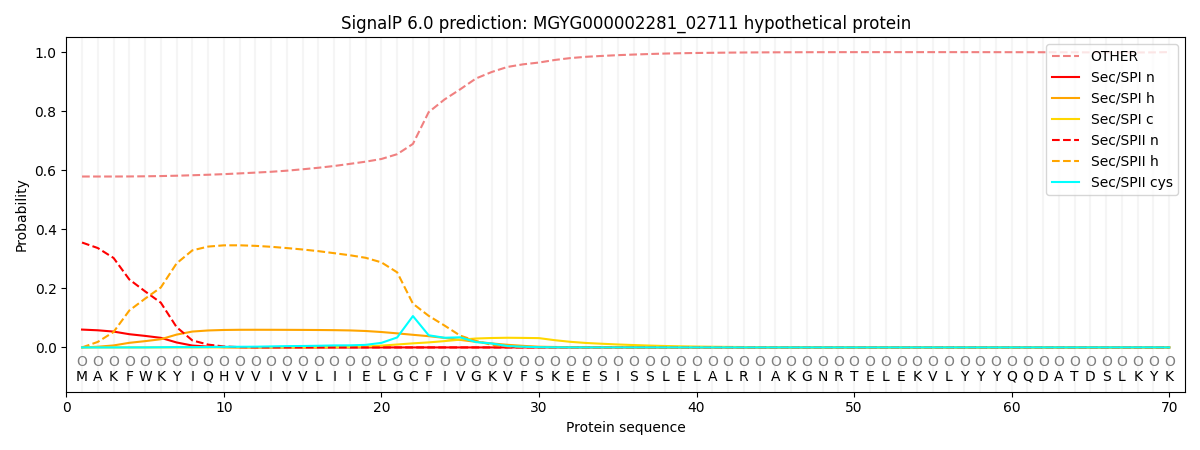
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.581657 | 0.056087 | 0.355897 | 0.000172 | 0.000138 | 0.006058 |