You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002300_01656
You are here: Home > Sequence: MGYG000002300_01656
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides cutis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides cutis | |||||||||||
| CAZyme ID | MGYG000002300_01656 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 411425; End: 413461 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 367 | 611 | 1.2e-54 | 0.3945945945945946 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06088 | KOW_RPL14 | 0.006 | 362 | 395 | 21 | 54 | KOW motif of Ribosomal Protein L14. RPL14 is a component of the large ribosomal subunit in both archaea and eukaryotes with KOW motif at its N terminal. KOW domain is known as an RNA-binding motif that is shared so far among some families of ribosomal proteins, the essential bacterial transcriptional elongation factor NusG, the eukaryotic chromatin elongation factor Spt5, the higher eukaryotic KIN17 proteins and Mtr4. Auto-antibodies to RPL14 in humans have been associated with systemic lupus erythematosus . Although RPL14 is well conserved, it is not found in all archaea, and therefore it is presumably not essential. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO71377.1 | 0.0 | 12 | 678 | 15 | 682 |
| ALL06947.1 | 0.0 | 21 | 676 | 24 | 679 |
| QBQ40168.1 | 0.0 | 21 | 674 | 21 | 674 |
| QMV69690.1 | 0.0 | 24 | 674 | 86 | 737 |
| QQT53092.1 | 0.0 | 24 | 674 | 86 | 737 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4TZ1_A | 7.21e-114 | 124 | 660 | 21 | 535 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4TZ3_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4TZ5_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4TZ5_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
| 4TYV_A | 7.65e-114 | 124 | 660 | 23 | 537 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4TYV_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E] |
| 4PEW_A | 1.03e-113 | 124 | 660 | 33 | 547 | Structureof sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4PEW_B Structure of sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 4PEX_A | 1.03e-113 | 124 | 660 | 33 | 547 | Structureof the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEX_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEY_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4PEZ_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4PF0_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4PF0_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2NFJ9 | 2.56e-113 | 124 | 660 | 77 | 591 | Exo-beta-1,3-glucanase OS=Streptomyces sp. (strain SirexAA-E / ActE) OX=862751 GN=SACTE_4363 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
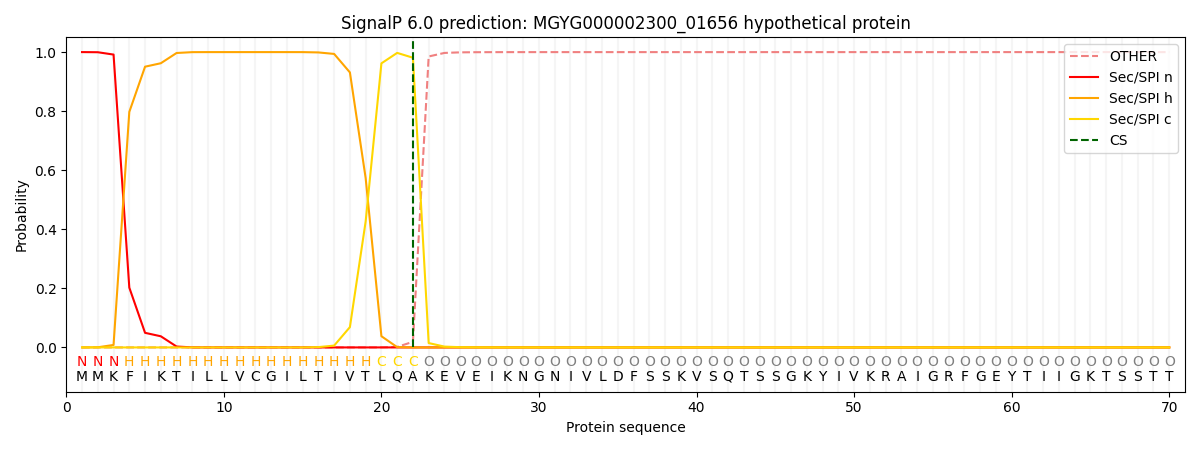
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000207 | 0.999165 | 0.000190 | 0.000140 | 0.000143 | 0.000136 |
