You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002312_01496
You are here: Home > Sequence: MGYG000002312_01496
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
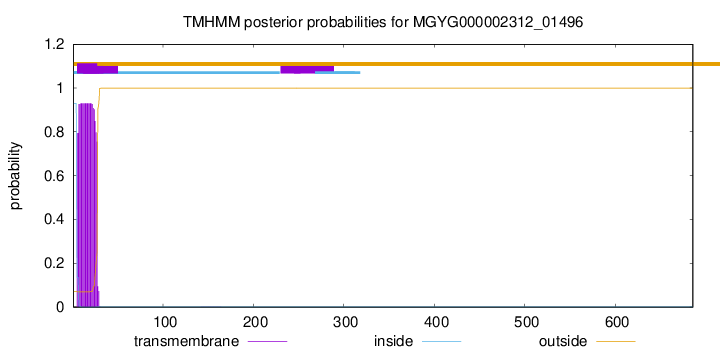
TMHMM annotations
Basic Information help
| Species | Blautia_A sp000285855 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia_A; Blautia_A sp000285855 | |||||||||||
| CAZyme ID | MGYG000002312_01496 | |||||||||||
| CAZy Family | CBM77 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10717; End: 12777 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PHA03247 | PHA03247 | 9.48e-23 | 435 | 594 | 2702 | 2875 | large tegument protein UL36; Provisional |
| NF033761 | gliding_GltJ | 1.29e-21 | 419 | 592 | 388 | 564 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| PHA03247 | PHA03247 | 6.34e-21 | 429 | 594 | 2708 | 2897 | large tegument protein UL36; Provisional |
| PHA03247 | PHA03247 | 7.59e-21 | 427 | 594 | 2725 | 2911 | large tegument protein UL36; Provisional |
| PHA03247 | PHA03247 | 5.11e-20 | 425 | 594 | 2729 | 2927 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZH69735.1 | 3.25e-112 | 59 | 408 | 115 | 469 |
| VCV20359.1 | 5.68e-15 | 588 | 665 | 1213 | 1289 |
| CBL07467.1 | 5.69e-15 | 588 | 665 | 1219 | 1295 |
| EEV00397.1 | 5.70e-15 | 588 | 665 | 1225 | 1301 |
| CBL14297.1 | 2.24e-14 | 588 | 665 | 1213 | 1289 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
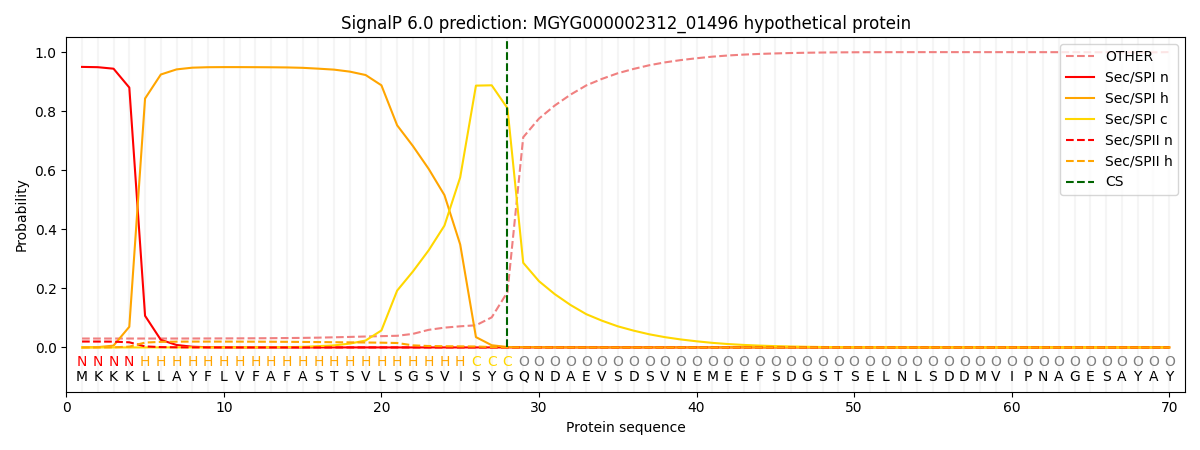
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.034264 | 0.943435 | 0.021341 | 0.000378 | 0.000272 | 0.000272 |