You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002327_02895
You are here: Home > Sequence: MGYG000002327_02895
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Dysosmobacter welbionis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Dysosmobacter; Dysosmobacter welbionis | |||||||||||
| CAZyme ID | MGYG000002327_02895 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 298; End: 1476 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 124 | 350 | 2.1e-45 | 0.9675925925925926 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 8.31e-67 | 65 | 384 | 1 | 315 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 1.14e-63 | 64 | 392 | 1 | 318 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK05337 | PRK05337 | 1.97e-30 | 93 | 349 | 27 | 277 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 9.27e-21 | 57 | 391 | 38 | 358 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCI59548.1 | 9.77e-277 | 1 | 392 | 1 | 392 |
| BCK84132.1 | 2.27e-178 | 1 | 392 | 1 | 396 |
| QNL44190.1 | 1.93e-175 | 1 | 392 | 1 | 383 |
| QUO36703.1 | 4.20e-175 | 39 | 392 | 37 | 398 |
| BAL01846.1 | 1.37e-155 | 1 | 392 | 1 | 403 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 1.73e-42 | 63 | 387 | 10 | 338 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 5BU9_A | 2.07e-36 | 62 | 386 | 3 | 337 | Crystalstructure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333],5BU9_B Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333] |
| 4YYF_A | 3.43e-36 | 61 | 381 | 36 | 354 | ChainA, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155],4YYF_B Chain B, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155],4YYF_C Chain C, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155] |
| 6GFV_A | 4.28e-35 | 62 | 386 | 14 | 337 | Mtuberculosis LpqI [Mycobacterium tuberculosis H37Rv],6GFV_B M tuberculosis LpqI [Mycobacterium tuberculosis H37Rv] |
| 4ZM6_A | 8.40e-33 | 66 | 386 | 9 | 337 | Aunique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432],4ZM6_B A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| L7N6B0 | 1.09e-35 | 3 | 386 | 5 | 380 | Beta-hexosaminidase LpqI OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lpqI PE=1 SV=1 |
| A0A0H3M1P5 | 2.90e-35 | 3 | 386 | 5 | 380 | Beta-hexosaminidase LpqI OS=Mycobacterium bovis (strain BCG / Pasteur 1173P2) OX=410289 GN=lpqI PE=3 SV=1 |
| P40406 | 1.83e-31 | 54 | 391 | 32 | 398 | Beta-hexosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagZ PE=1 SV=1 |
| B2FPW9 | 5.85e-26 | 92 | 371 | 24 | 296 | Beta-hexosaminidase OS=Stenotrophomonas maltophilia (strain K279a) OX=522373 GN=nagZ PE=3 SV=1 |
| B4SRK3 | 1.11e-25 | 92 | 371 | 24 | 296 | Beta-hexosaminidase OS=Stenotrophomonas maltophilia (strain R551-3) OX=391008 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
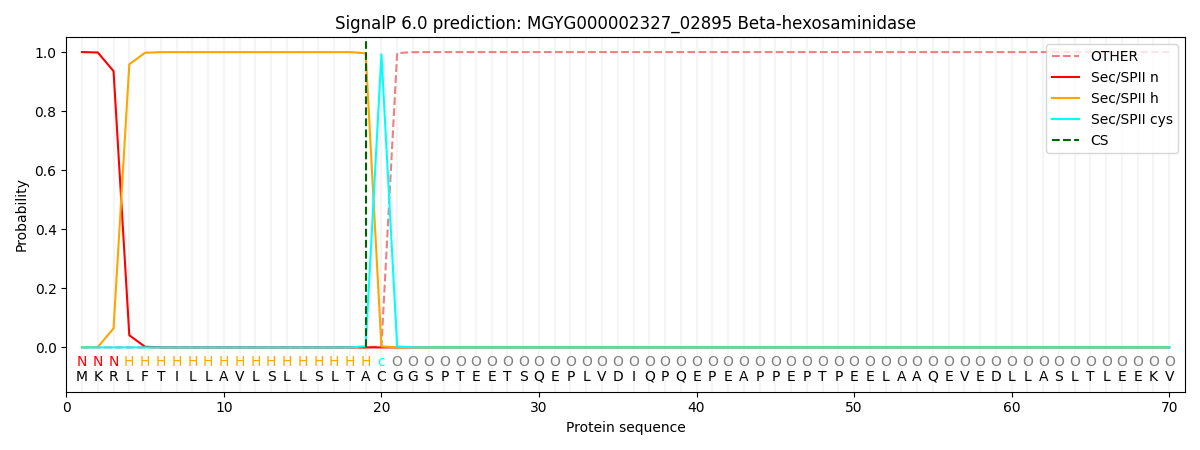
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000063 | 0.000000 | 0.000000 | 0.000000 |
