You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002335_01202
You are here: Home > Sequence: MGYG000002335_01202
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Yersinia enterocolitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Yersinia; Yersinia enterocolitica | |||||||||||
| CAZyme ID | MGYG000002335_01202 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | Glutamate/aspartate import solute-binding protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1320226; End: 1321140 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10797 | PRK10797 | 0.0 | 1 | 304 | 1 | 302 | glutamate and aspartate transporter subunit; Provisional |
| cd13688 | PBP2_GltI_DEBP | 7.45e-114 | 35 | 272 | 1 | 238 | Substrate-binding domain of ABC aspartate-glutamate transporter; the type 2 periplasmic binding protein fold. This subfamily represents the periplasmic-binding protein component of ABC transporter specific for carboxylic amino acids, including GtlI from Escherichia coli. The aspartate-glutamate binding domain belongs to the type 2 periplasmic binding protein fold superfamily (PBP2), whose many members are involved in chemotaxis and uptake of nutrients and other small molecules from the extracellular space as a primary receptor. The PBP2 proteins are typically comprised of two globular subdomains connected by a flexible hinge and bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. After binding their specific ligand with high affinity, they can interact with a cognate membrane transport complex comprised of two integral membrane domains and two receptor cytoplasmically-located ATPase domains. This interaction triggers the ligand translocation across the cytoplasmic membrane energized by ATP hydrolysis. |
| cd01000 | PBP2_Cys_DEBP_like | 1.61e-82 | 35 | 272 | 1 | 228 | Substrate-binding domain of cysteine- and aspartate/glutamate-binding proteins; the type 2 periplasmic-binding protein fold. This family comprises of the periplasmic-binding protein component of ABC transporters specific for cysteine and carboxylic amino acids, as well as their closely related proteins. The cysteine and aspartate-glutamate binding domains belong to the type 2 periplasmic binding protein fold superfamily (PBP2), whose many members are involved in chemotaxis and uptake of nutrients and other small molecules from the extracellular space as a primary receptor. The PBP2 proteins are typically comprised of two globular subdomains connected by a flexible hinge and bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. After binding their specific ligand with high affinity, they can interact with a cognate membrane transport complex comprised of two integral membrane domains and two receptor cytoplasmically-located ATPase domains. This interaction triggers the ligand translocation across the cytoplasmic membrane energized by ATP hydrolysis. |
| cd13689 | PBP2_BsGlnH | 4.33e-63 | 35 | 273 | 1 | 229 | Substrate binding domain of ABC glutamine transporter from Bacillus subtilis; the type 2 periplasmic-bindig protein fold. This group includes periplasmic glutamine-binding domain GlnP from Bacillus subtilis and its related proteins. The GlnP domain belongs to the type 2 periplasmic binding protein fold superfamily (PBP2), whose many members are involved in chemotaxis and uptake of nutrients and other small molecules from the extracellular space as a primary receptor. The PBP2 proteins are typically comprised of two globular subdomains connected by a flexible hinge and bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. After binding their specific ligand with high affinity, they can interact with a cognate membrane transport complex comprised of two integral membrane domains and two receptor cytoplasmically-located ATPase domains. This interaction triggers the ligand translocation across the cytoplasmic membrane energized by ATP hydrolysis. |
| smart00062 | PBPb | 1.21e-54 | 43 | 272 | 1 | 219 | Bacterial periplasmic substrate-binding proteins. bacterial proteins, eukaryotic ones are in PBPe |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QXX81055.1 | 5.23e-171 | 1 | 304 | 1 | 297 |
| QWJ92671.1 | 3.02e-170 | 1 | 304 | 1 | 297 |
| QLQ94799.1 | 3.02e-170 | 1 | 304 | 1 | 297 |
| QQE94206.1 | 3.02e-170 | 1 | 304 | 1 | 297 |
| BBV05439.1 | 8.65e-170 | 1 | 304 | 1 | 297 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VHA_A | 1.33e-172 | 30 | 304 | 5 | 279 | DEBP[Shigella flexneri],2VHA_B DEBP [Shigella flexneri] |
| 2IA4_A | 3.31e-166 | 30 | 304 | 5 | 279 | Crystalstructure of Novel amino acid binding protein from Shigella flexneri [Shigella flexneri 2a str. 301],2IA4_B Crystal structure of Novel amino acid binding protein from Shigella flexneri [Shigella flexneri 2a str. 301] |
| 5EYF_A | 7.27e-27 | 29 | 274 | 2 | 238 | CrystalStructure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate [Enterococcus faecium DO],5EYF_B Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate [Enterococcus faecium DO] |
| 3K4U_A | 6.42e-14 | 42 | 272 | 5 | 227 | CRYSTALSTRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine [Wolinella succinogenes],3K4U_B CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine [Wolinella succinogenes],3K4U_C CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine [Wolinella succinogenes],3K4U_D CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine [Wolinella succinogenes],3K4U_E CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine [Wolinella succinogenes],3K4U_F CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine [Wolinella succinogenes] |
| 1XT8_A | 1.50e-13 | 7 | 273 | 9 | 265 | ChainA, putative amino-acid transporter periplasmic solute-binding protein [Campylobacter jejuni],1XT8_B Chain B, putative amino-acid transporter periplasmic solute-binding protein [Campylobacter jejuni] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ZF60 | 2.50e-178 | 1 | 304 | 1 | 302 | Glutamate/aspartate import solute-binding protein OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=gltI PE=3 SV=3 |
| P37902 | 5.60e-175 | 1 | 304 | 1 | 302 | Glutamate/aspartate import solute-binding protein OS=Escherichia coli (strain K12) OX=83333 GN=gltI PE=1 SV=2 |
| Q9I402 | 1.01e-111 | 29 | 302 | 24 | 298 | L-glutamate/L-aspartate-binding protein OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=PA1342 PE=1 SV=1 |
| O34563 | 7.23e-24 | 10 | 273 | 16 | 268 | ABC transporter glutamine-binding protein GlnH OS=Bacillus subtilis (strain 168) OX=224308 GN=glnH PE=2 SV=1 |
| P27676 | 1.54e-21 | 17 | 254 | 31 | 262 | Glutamine-binding protein OS=Geobacillus stearothermophilus OX=1422 GN=glnH PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
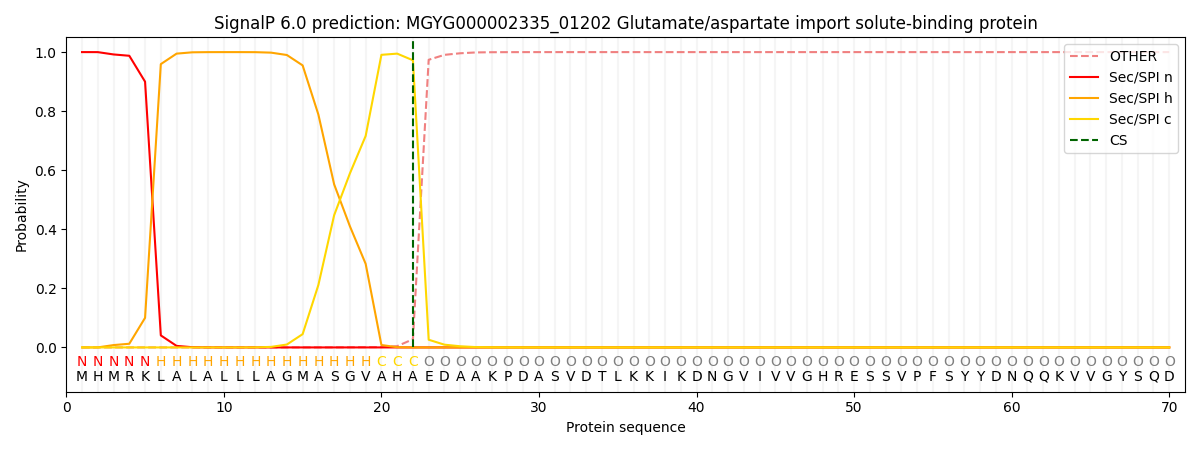
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000314 | 0.998923 | 0.000232 | 0.000203 | 0.000170 | 0.000148 |
