You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002357_00345
You are here: Home > Sequence: MGYG000002357_00345
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus licheniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus licheniformis | |||||||||||
| CAZyme ID | MGYG000002357_00345 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | Chitodextrinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 317875; End: 319956 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 68 | 521 | 1.3e-71 | 0.9425675675675675 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3325 | ChiA | 1.00e-136 | 41 | 521 | 11 | 425 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
| cd06548 | GH18_chitinase | 1.45e-98 | 70 | 519 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| smart00636 | Glyco_18 | 4.05e-93 | 69 | 519 | 1 | 334 | Glyco_18 domain. |
| pfam06483 | ChiC | 3.35e-67 | 532 | 692 | 1 | 161 | Chitinase C. This ~170 aa region is found at the C-terminus of pfam00704. |
| pfam00704 | Glyco_hydro_18 | 1.14e-66 | 69 | 519 | 1 | 307 | Glycosyl hydrolases family 18. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAW36031.1 | 0.0 | 1 | 693 | 1 | 693 |
| QQA72206.1 | 0.0 | 1 | 693 | 1 | 693 |
| QSV40254.1 | 0.0 | 1 | 693 | 1 | 693 |
| AKQ71443.1 | 0.0 | 1 | 693 | 1 | 693 |
| AWV39225.1 | 0.0 | 1 | 693 | 1 | 693 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4TXG_A | 8.45e-301 | 35 | 691 | 121 | 781 | CrystalStructure of a Family GH18 Chitinase from Chromobacterium violaceum [Chromobacterium violaceum ATCC 12472] |
| 1ITX_A | 6.38e-47 | 69 | 519 | 13 | 406 | ChainA, Glycosyl Hydrolase [Niallia circulans] |
| 6BT9_A | 3.21e-46 | 68 | 525 | 37 | 454 | ChitinaseChiA74 from Bacillus thuringiensis [Bacillus thuringiensis],6BT9_B Chitinase ChiA74 from Bacillus thuringiensis [Bacillus thuringiensis] |
| 6KST_A | 2.63e-45 | 69 | 519 | 8 | 357 | Crystalstructure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KST_B Crystal structure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KXL_A Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXL_B Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
| 6KXM_A | 1.29e-44 | 69 | 519 | 8 | 357 | Crystalstructure of D157N mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXM_B Crystal structure of D157N mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96156 | 1.25e-256 | 56 | 691 | 308 | 964 | Chitodextrinase OS=Vibrio furnissii OX=29494 GN=endo I PE=1 SV=1 |
| P20533 | 1.45e-44 | 2 | 519 | 11 | 438 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P32470 | 7.84e-29 | 53 | 517 | 25 | 381 | Chitinase 1 OS=Aphanocladium album OX=12942 GN=CHI1 PE=1 SV=2 |
| P11797 | 8.28e-29 | 68 | 519 | 5 | 408 | Chitinase B OS=Serratia marcescens OX=615 GN=chiB PE=1 SV=1 |
| P32823 | 7.12e-28 | 57 | 519 | 146 | 573 | Chitinase A OS=Pseudoalteromonas piscicida OX=43662 GN=chiA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
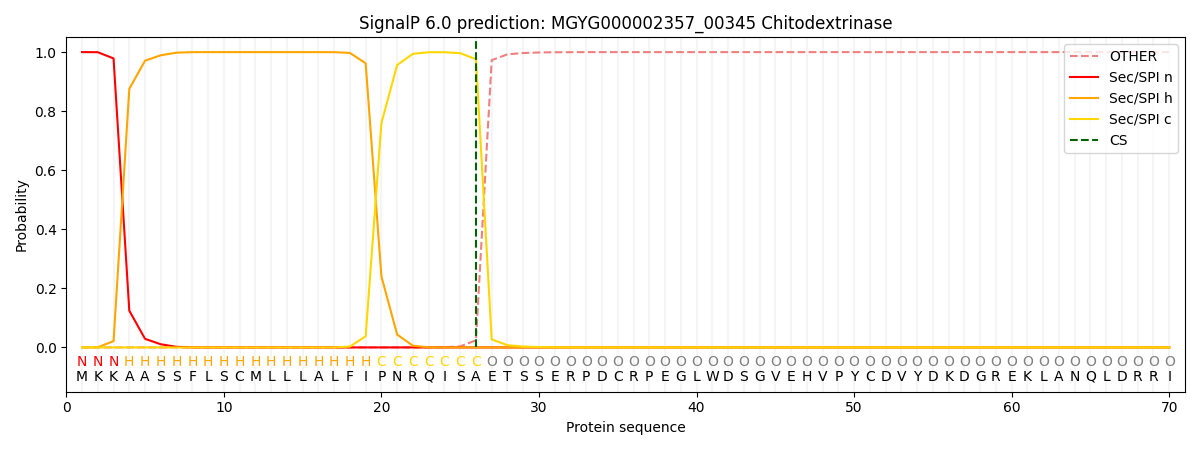
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000198 | 0.999153 | 0.000180 | 0.000158 | 0.000142 | 0.000130 |
