You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002363_03934
You are here: Home > Sequence: MGYG000002363_03934
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Raoultella ornithinolytica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Raoultella; Raoultella ornithinolytica | |||||||||||
| CAZyme ID | MGYG000002363_03934 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Membrane-bound lytic murein transglycosylase C | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 106167; End: 107252 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 209 | 356 | 8.2e-22 | 0.7851851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK11671 | mltC | 0.0 | 2 | 361 | 1 | 359 | membrane-bound lytic murein transglycosylase MltC. |
| cd16893 | LT_MltC_MltE | 1.02e-98 | 197 | 356 | 2 | 162 | membrane-bound lytic murein transglycosylases MltC and MltE, and similar proteins. MltC and MltE are periplasmic, outer membrane attached lytic transglycosylases (LTs), which cleave beta-1,4-glycosidic bonds joining N-acetylmuramic acid and N-acetylglucosamine in the cell wall peptidoglycan, yielding 1,6-anhydromuropeptides. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda |
| pfam11873 | DUF3393 | 4.37e-98 | 32 | 193 | 1 | 161 | Domain of unknown function (DUF3393). This domain is functionally uncharacterized. This domain is found in bacteria. This presumed domain is typically between 188 to 206 amino acids in length. This domain is found associated with pfam01464. |
| COG0741 | MltE | 1.56e-54 | 49 | 356 | 1 | 296 | Soluble lytic murein transglycosylase and related regulatory proteins (some contain LysM/invasin domains) [Cell wall/membrane/envelope biogenesis]. |
| PRK15470 | emtA | 2.09e-43 | 189 | 354 | 34 | 199 | membrane-bound lytic murein transglycosylase EmtA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALQ44895.1 | 1.36e-265 | 1 | 361 | 1 | 361 |
| BBQ87667.1 | 1.36e-265 | 1 | 361 | 1 | 361 |
| VEB66145.1 | 1.36e-265 | 1 | 361 | 1 | 361 |
| ASI59152.1 | 1.36e-265 | 1 | 361 | 1 | 361 |
| QHW67006.1 | 1.36e-265 | 1 | 361 | 1 | 361 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4C5F_A | 1.23e-221 | 23 | 361 | 3 | 341 | Structureof Lytic Transglycosylase MltC from Escherichia coli at 2.3 A resolution. [Escherichia coli],4C5F_B Structure of Lytic Transglycosylase MltC from Escherichia coli at 2.3 A resolution. [Escherichia coli] |
| 4CFO_A | 3.52e-221 | 23 | 361 | 3 | 341 | Structureof Lytic Transglycosylase MltC from Escherichia coli in complex with tetrasaccharide at 2.9 A resolution. [Escherichia coli],4CFO_B Structure of Lytic Transglycosylase MltC from Escherichia coli in complex with tetrasaccharide at 2.9 A resolution. [Escherichia coli],4CFP_A Crystal structure of MltC in complex with tetrasaccharide at 2.15 A resolution [Escherichia coli],4CFP_B Crystal structure of MltC in complex with tetrasaccharide at 2.15 A resolution [Escherichia coli],4CHX_A Crystal structure of MltC in complex with disaccharide pentapeptide DHl89 [Escherichia coli],4CHX_B Crystal structure of MltC in complex with disaccharide pentapeptide DHl89 [Escherichia coli] |
| 6GI4_B | 4.77e-35 | 189 | 354 | 17 | 182 | Structureof Lytic Transglycosylase MltE mutant S75A from E.coli [Escherichia coli] |
| 2Y8P_A | 1.31e-34 | 189 | 354 | 17 | 182 | CrystalStructure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli [Escherichia coli K-12],2Y8P_B Crystal Structure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli [Escherichia coli K-12] |
| 3T36_A | 1.68e-34 | 189 | 354 | 34 | 199 | Crystalstructure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],3T36_B Crystal structure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],3T36_C Crystal structure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],3T36_D Crystal structure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],3T36_E Crystal structure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],4HJV_A Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12],4HJV_B Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12],4HJV_C Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12],4HJV_D Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12],4HJV_E Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B5XU88 | 7.25e-248 | 2 | 361 | 1 | 360 | Membrane-bound lytic murein transglycosylase C OS=Klebsiella pneumoniae (strain 342) OX=507522 GN=mltC PE=3 SV=1 |
| A6TDX1 | 2.95e-247 | 2 | 361 | 1 | 360 | Membrane-bound lytic murein transglycosylase C OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=mltC PE=3 SV=2 |
| A8API2 | 1.86e-234 | 2 | 361 | 1 | 360 | Membrane-bound lytic murein transglycosylase C OS=Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696) OX=290338 GN=mltC PE=3 SV=2 |
| B7LPT3 | 3.46e-232 | 2 | 361 | 1 | 359 | Membrane-bound lytic murein transglycosylase C OS=Escherichia fergusonii (strain ATCC 35469 / DSM 13698 / CCUG 18766 / IAM 14443 / JCM 21226 / LMG 7866 / NBRC 102419 / NCTC 12128 / CDC 0568-73) OX=585054 GN=mltC PE=3 SV=1 |
| P0C066 | 9.92e-232 | 2 | 361 | 1 | 359 | Membrane-bound lytic murein transglycosylase C OS=Escherichia coli (strain K12) OX=83333 GN=mltC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
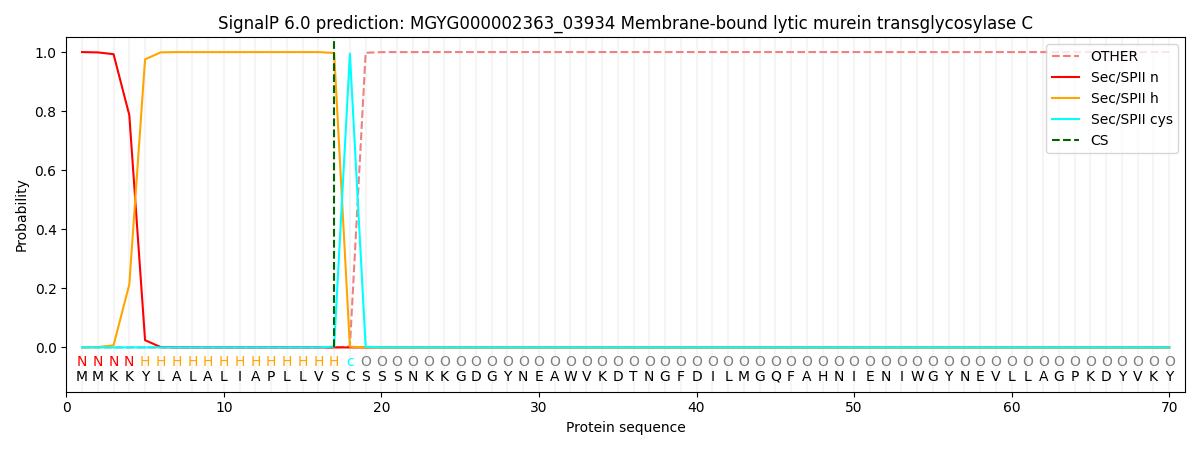
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000076 | 0.000000 | 0.000000 | 0.000000 |
