You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002369_02444
You are here: Home > Sequence: MGYG000002369_02444
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
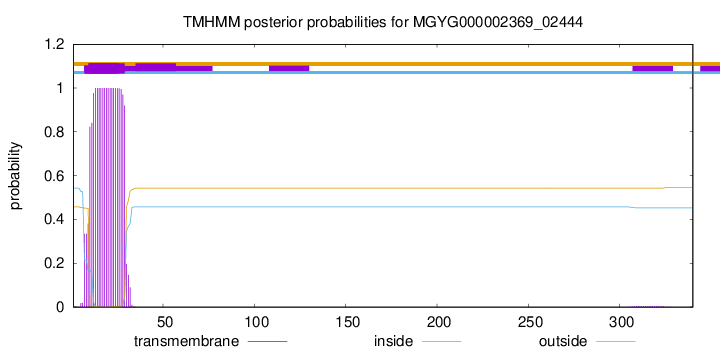
TMHMM annotations
Basic Information help
| Species | Clostridioides difficile | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Clostridioides; Clostridioides difficile | |||||||||||
| CAZyme ID | MGYG000002369_02444 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase H | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2671294; End: 2672316 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 56 | 314 | 9.6e-103 | 0.9847328244274809 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.67e-50 | 64 | 312 | 29 | 266 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 5.23e-34 | 16 | 320 | 25 | 368 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVB34576.1 | 6.55e-251 | 1 | 340 | 1 | 340 |
| AVB54202.1 | 6.55e-251 | 1 | 340 | 1 | 340 |
| AVB57828.1 | 6.55e-251 | 1 | 340 | 1 | 340 |
| AYD25742.1 | 6.55e-251 | 1 | 340 | 1 | 340 |
| QGS41366.1 | 6.55e-251 | 1 | 340 | 1 | 340 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UJE_A | 1.54e-225 | 30 | 340 | 2 | 312 | ChainA, Endoglucanase [Clostridioides difficile],6UJF_A Chain A, Endoglucanase [Clostridioides difficile] |
| 3AMC_A | 4.84e-90 | 40 | 337 | 14 | 311 | Crystalstructures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMC_B Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMD_A Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_B Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_C Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_D Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3MMU_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_E Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_F Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_G Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_H Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima] |
| 3AZR_A | 3.90e-89 | 40 | 337 | 14 | 311 | DiverseSubstrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose [Thermotoga maritima MSB8],3AZR_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose [Thermotoga maritima MSB8],3AZS_A Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose [Thermotoga maritima MSB8],3AZS_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose [Thermotoga maritima MSB8],3AZT_A Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_C Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_D Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8] |
| 3AMG_A | 3.90e-89 | 40 | 337 | 14 | 311 | Crystalstructures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8],3AMG_B Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8] |
| 3AOF_A | 3.56e-87 | 40 | 337 | 14 | 311 | Crystalstructures of Thermotoga maritima Cel5A in complex with Mannotriose substrate [Thermotoga maritima MSB8],3AOF_B Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P25472 | 3.46e-56 | 13 | 337 | 13 | 328 | Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
| P16218 | 2.28e-55 | 37 | 336 | 335 | 628 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
| P28623 | 1.58e-30 | 37 | 338 | 49 | 370 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P10477 | 4.12e-28 | 34 | 313 | 59 | 350 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| O08342 | 1.70e-26 | 35 | 338 | 44 | 397 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
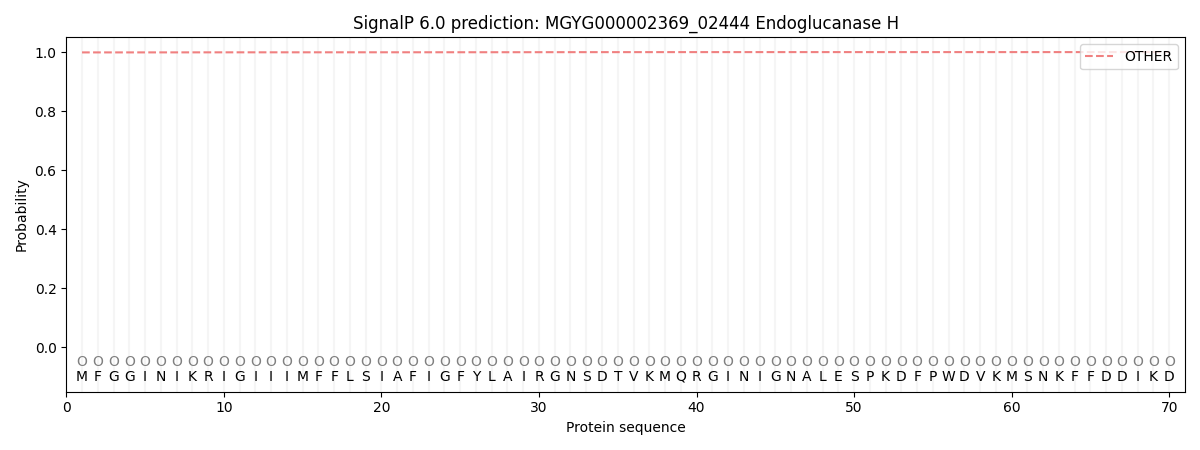
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999028 | 0.000813 | 0.000090 | 0.000004 | 0.000002 | 0.000080 |