You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002372_02356
You are here: Home > Sequence: MGYG000002372_02356
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
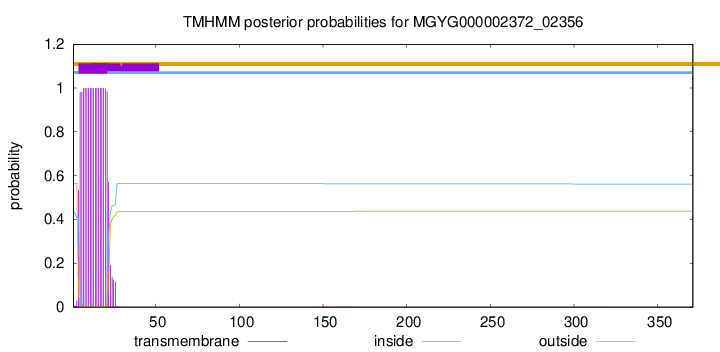
TMHMM annotations
Basic Information help
| Species | Clostridium_P perfringens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_P; Clostridium_P perfringens | |||||||||||
| CAZyme ID | MGYG000002372_02356 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2581010; End: 2582125 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 46 | 365 | 1.4e-130 | 0.9937694704049844 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 3.39e-07 | 70 | 165 | 17 | 129 | Glycosyl hydrolases family 8. |
| COG3405 | BcsZ | 7.27e-06 | 55 | 169 | 37 | 149 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMN33485.1 | 7.38e-275 | 1 | 371 | 1 | 371 |
| AOY54757.1 | 7.38e-275 | 1 | 371 | 1 | 371 |
| ABG82272.1 | 1.05e-274 | 1 | 371 | 1 | 371 |
| QUD72463.1 | 1.05e-274 | 1 | 371 | 1 | 371 |
| ALG49594.1 | 1.05e-274 | 1 | 371 | 1 | 371 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3REN_A | 1.81e-237 | 43 | 371 | 22 | 350 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
| 1CEM_A | 7.02e-09 | 49 | 225 | 33 | 216 | ChainA, CELLULASE CELA (1,4-BETA-D-GLUCAN-GLUCANOHYDROLASE) [Acetivibrio thermocellus],1IS9_A Chain A, endoglucanase A [Acetivibrio thermocellus] |
| 1KWF_A | 3.94e-08 | 49 | 225 | 33 | 216 | ChainA, Endoglucanase A [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 8.15e-25 | 55 | 368 | 41 | 354 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
| A3DC29 | 1.17e-07 | 49 | 225 | 65 | 248 | Endoglucanase A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
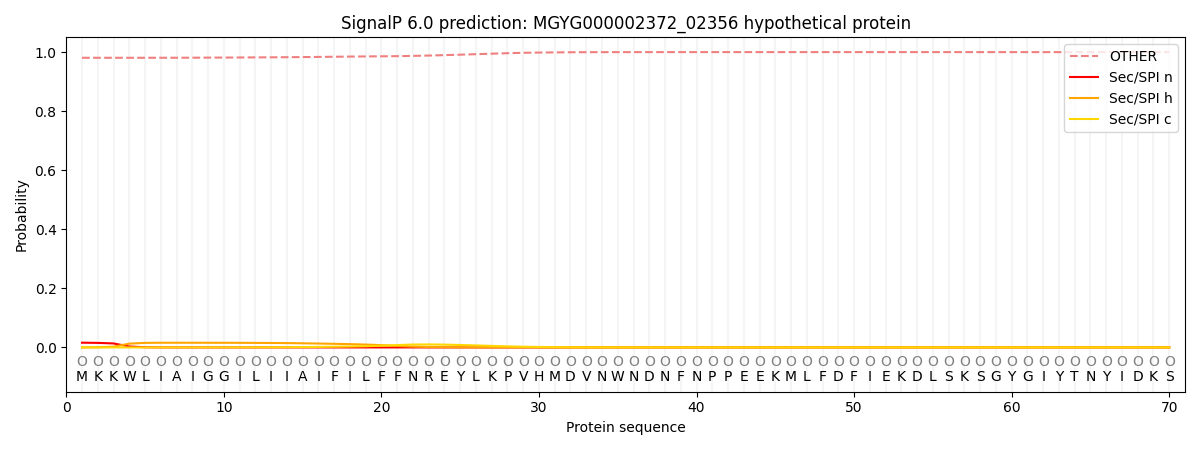
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.981700 | 0.014729 | 0.003375 | 0.000040 | 0.000034 | 0.000145 |