You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002376_02642
You are here: Home > Sequence: MGYG000002376_02642
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
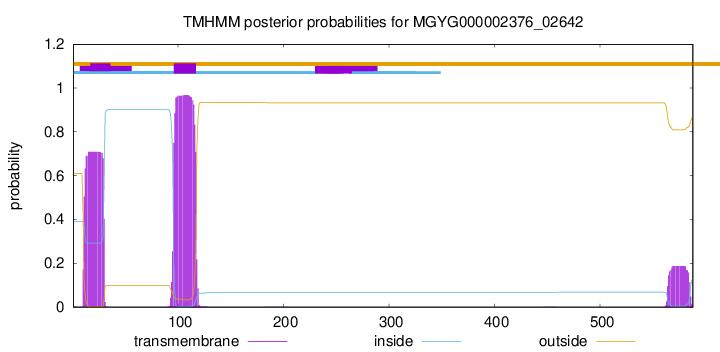
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium_B sp000765215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium_B; Lachnoclostridium_B sp000765215 | |||||||||||
| CAZyme ID | MGYG000002376_02642 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19426; End: 21192 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 299 | 460 | 8.3e-29 | 0.9529411764705882 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02397 | Bac_transf | 1.47e-74 | 91 | 262 | 1 | 173 | Bacterial sugar transferase. This Pfam family represents a conserved region from a number of different bacterial sugar transferases, involved in diverse biosynthesis pathways. |
| COG2148 | WcaJ | 4.61e-71 | 62 | 274 | 6 | 226 | Sugar transferase involved in LPS biosynthesis (colanic, teichoic acid) [Cell wall/membrane/envelope biogenesis]. |
| TIGR03025 | EPS_sugtrans | 4.50e-56 | 87 | 259 | 254 | 429 | exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase. Members of this family are generally found near other genes involved in the biosynthesis of a variety of exopolysaccharides. These proteins consist of two fused domains, an N-terminal hydrophobic domain of generally low conservation and a highly conserved C-terminal sugar transferase domain (pfam02397). Characterized and partially characterized members of this subfamily include Salmonella WbaP (originally RfbP), E. coli WcaJ, Methylobacillus EpsB, Xanthomonas GumD, Vibrio CpsA, Erwinia AmsG, Group B Streptococcus CpsE (originally CpsD), and Streptococcus suis Cps2E. Each of these is believed to act in transferring the sugar from, for instance, UDP-glucose or UDP-galactose, to a lipid carrier such as undecaprenyl phosphate as the first (priming) step in the synthesis of an oligosaccharide "block". This function is encoded in the C-terminal domain. The liposaccharide is believed to be subsequently transferred through a "flippase" function from the cytoplasmic to the periplasmic face of the inner membrane by the N-terminal domain. Certain closely related transferase enzymes, such as Sinorhizobium ExoY and Lactococcus EpsD, lack the N-terminal domain and are not found by this model. |
| PRK10124 | PRK10124 | 6.63e-34 | 91 | 259 | 273 | 447 | putative UDP-glucose lipid carrier transferase; Provisional |
| pfam00535 | Glycos_transf_2 | 7.15e-31 | 299 | 460 | 1 | 161 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUO31262.1 | 3.20e-132 | 1 | 474 | 1 | 498 |
| QNU03878.1 | 3.94e-100 | 271 | 584 | 3 | 313 |
| QMW80818.1 | 2.33e-78 | 90 | 284 | 34 | 228 |
| QIB56408.1 | 2.33e-78 | 90 | 284 | 34 | 228 |
| AWG44220.1 | 1.79e-67 | 298 | 588 | 5 | 302 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5W7L_A | 1.47e-29 | 86 | 284 | 6 | 200 | Structureof Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826],5W7L_B Structure of Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826] |
| 1H7L_A | 2.50e-15 | 298 | 431 | 3 | 143 | dTDP-MAGNESIUMCOMPLEX OF SPSA FROM BACILLUS SUBTILIS [Bacillus subtilis],1H7Q_A dTDP-MANGANESE COMPLEX OF SPSA FROM BACILLUS SUBTILIS [Bacillus subtilis],1QG8_A Native (Magnesium-Containing) Spsa From Bacillus Subtilis [Bacillus subtilis],1QGQ_A Udp-manganese Complex Of Spsa From Bacillus Subtilis [Bacillus subtilis],1QGS_A Udp-Magnesium Complex Of Spsa From Bacillus Subtilis [Bacillus subtilis] |
| 5HEA_A | 3.84e-13 | 297 | 424 | 6 | 129 | CgTstructure in hexamer [Streptococcus parasanguinis FW213],5HEA_B CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEA_C CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEC_A CgT structure in dimer [Streptococcus parasanguinis FW213],5HEC_B CgT structure in dimer [Streptococcus parasanguinis FW213] |
| 6P61_A | 4.39e-08 | 298 | 429 | 15 | 143 | Structureof a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_B Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_C Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_D Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197] |
| 5TZE_C | 1.13e-06 | 299 | 410 | 4 | 114 | Crystalstructure of S. aureus TarS in complex with UDP-GlcNAc [Staphylococcus aureus],5TZE_E Crystal structure of S. aureus TarS in complex with UDP-GlcNAc [Staphylococcus aureus],5TZI_C Crystal structure of S. aureus TarS 1-349 [Staphylococcus aureus],5TZJ_A Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc [Staphylococcus aureus],5TZJ_C Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc [Staphylococcus aureus],5TZK_C Crystal structure of S. aureus TarS 1-349 in complex with UDP [Staphylococcus aureus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P71241 | 2.76e-30 | 90 | 259 | 273 | 448 | UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferase OS=Escherichia coli (strain K12) OX=83333 GN=wcaJ PE=1 SV=2 |
| Q0P9D0 | 4.10e-30 | 86 | 283 | 2 | 195 | Undecaprenyl phosphate N,N'-diacetylbacillosamine 1-phosphate transferase OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) OX=192222 GN=pglC PE=1 SV=1 |
| Q44576 | 1.63e-27 | 91 | 259 | 338 | 516 | Undecaprenyl-phosphate glucose phosphotransferase OS=Komagataeibacter xylinus OX=28448 GN=aceA PE=3 SV=1 |
| Q56770 | 2.59e-27 | 91 | 270 | 293 | 480 | UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferase OS=Xanthomonas campestris pv. campestris OX=340 GN=gumD PE=1 SV=1 |
| P10498 | 4.00e-24 | 90 | 265 | 5 | 190 | Exopolysaccharide production protein PSS OS=Rhizobium leguminosarum bv. phaseoli OX=385 GN=pss PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
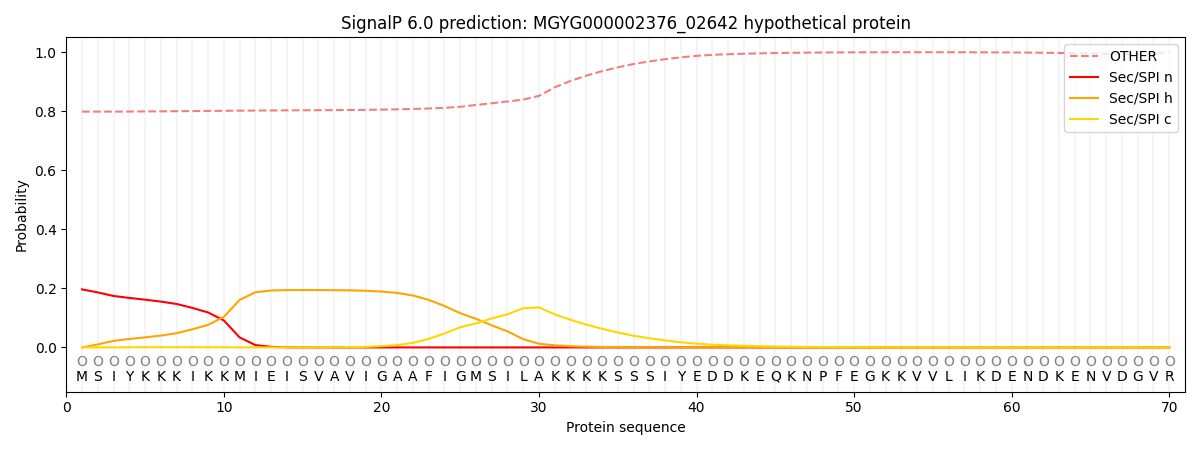
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.808793 | 0.181956 | 0.002501 | 0.001072 | 0.000596 | 0.005094 |