You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002399_00965
You are here: Home > Sequence: MGYG000002399_00965
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus hominis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus hominis | |||||||||||
| CAZyme ID | MGYG000002399_00965 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | Bifunctional autolysin | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 216119; End: 220159 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 1193 | 1319 | 6.9e-21 | 0.8984375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.36e-71 | 1106 | 1345 | 18 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| COG5632 | CwlA | 1.00e-54 | 366 | 670 | 1 | 300 | N-acetylmuramoyl-L-alanine amidase CwlA [Cell wall/membrane/envelope biogenesis]. |
| smart00047 | LYZ2 | 6.32e-30 | 1182 | 1339 | 2 | 146 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| smart00644 | Ami_2 | 1.20e-26 | 380 | 508 | 1 | 126 | Ami_2 domain. |
| cd06583 | PGRP | 1.45e-23 | 381 | 511 | 1 | 126 | Peptidoglycan recognition proteins (PGRPs) are pattern recognition receptors that bind, and in certain cases, hydrolyze peptidoglycans (PGNs) of bacterial cell walls. PGRPs have been divided into three classes: short PGRPs (PGRP-S), that are small (20 kDa) extracellular proteins; intermediate PGRPs (PGRP-I) that are 40-45 kDa and are predicted to be transmembrane proteins; and long PGRPs (PGRP-L), up to 90 kDa, which may be either intracellular or transmembrane. Several structures of PGRPs are known in insects and mammals, some bound with substrates like Muramyl Tripeptide (MTP) or Tracheal Cytotoxin (TCT). The substrate binding site is conserved in PGRP-LCx, PGRP-LE, and PGRP-Ialpha proteins. This family includes Zn-dependent N-Acetylmuramoyl-L-alanine Amidase, EC:3.5.1.28. This enzyme cleaves the amide bond between N-acetylmuramoyl and L-amino acids, preferentially D-lactyl-L-Ala, in bacterial cell walls. The structure for the bacteriophage T7 lysozyme shows that two of the conserved histidines and a cysteine are zinc binding residues. Site-directed mutagenesis of T7 lysozyme indicates that two conserved residues, a Tyr and a Lys, are important for amidase activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIY37350.1 | 0.0 | 1 | 1346 | 1 | 1346 |
| QDW87235.1 | 0.0 | 1 | 1346 | 1 | 1346 |
| AUW62106.1 | 0.0 | 1 | 1346 | 1 | 1346 |
| AVI05602.1 | 0.0 | 1 | 1346 | 1 | 1346 |
| AUJ52718.1 | 0.0 | 1 | 1346 | 1 | 1343 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4KNK_A | 5.93e-123 | 335 | 544 | 16 | 225 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNK_B Chain B, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_A Chain A, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_B Chain B, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_C Chain C, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_D Chain D, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 3LAT_A | 2.89e-119 | 334 | 541 | 6 | 213 | Crystalstructure of Staphylococcus peptidoglycan hydrolase AmiE [Staphylococcus epidermidis],3LAT_B Crystal structure of Staphylococcus peptidoglycan hydrolase AmiE [Staphylococcus epidermidis] |
| 6FXO_A | 3.55e-95 | 1103 | 1342 | 3 | 241 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4EPC_A | 4.58e-84 | 542 | 862 | 3 | 327 | Crystalstructure of Autolysin repeat domains from Staphylococcus epidermidis [Staphylococcus epidermidis] |
| 7KWI_A | 1.20e-60 | 558 | 707 | 1 | 150 | ChainA, Bifunctional autolysin [Staphylococcus epidermidis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5HQB9 | 0.0 | 1 | 1344 | 1 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q6GI31 | 0.0 | 1 | 1342 | 1 | 1254 | Bifunctional autolysin OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=atl PE=3 SV=1 |
| Q6GAG0 | 0.0 | 1 | 1342 | 1 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q8CPQ1 | 0.0 | 1 | 1344 | 1 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q5HH31 | 0.0 | 1 | 1342 | 1 | 1253 | Bifunctional autolysin OS=Staphylococcus aureus (strain COL) OX=93062 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
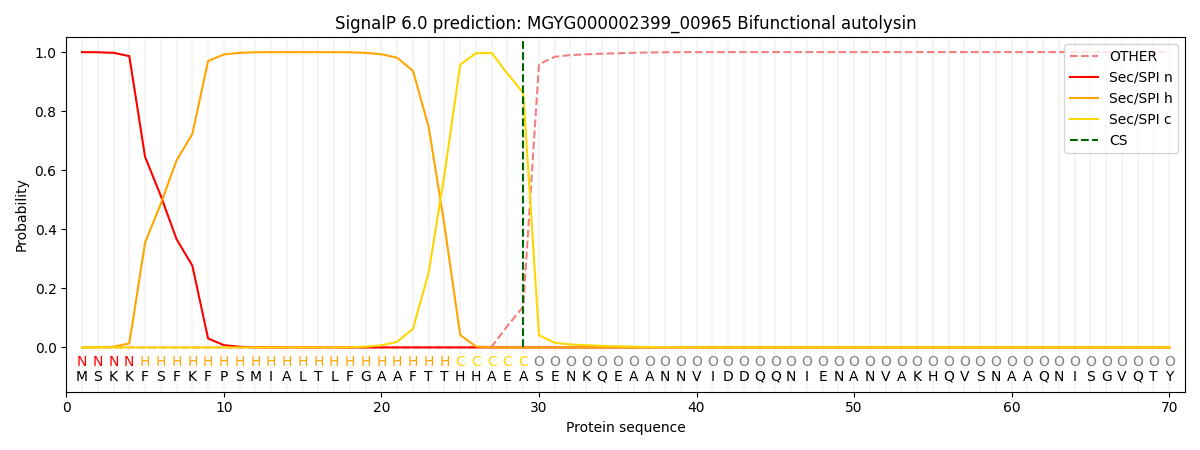
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000353 | 0.998707 | 0.000388 | 0.000187 | 0.000162 | 0.000148 |
