You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002403_05608
You are here: Home > Sequence: MGYG000002403_05608
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
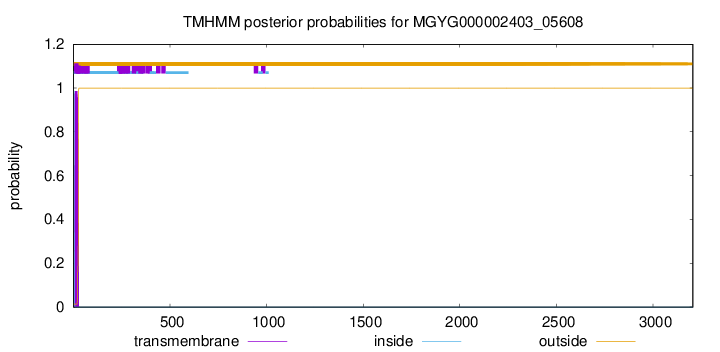
TMHMM annotations
Basic Information help
| Species | Robinsoniella peoriensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella peoriensis | |||||||||||
| CAZyme ID | MGYG000002403_05608 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 259957; End: 269577 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 1670 | 2221 | 5.5e-173 | 0.948905109489051 |
| GH2 | 33 | 625 | 2e-89 | 0.6170212765957447 |
| CBM51 | 1440 | 1585 | 1.6e-37 | 0.9925373134328358 |
| CBM32 | 1027 | 1155 | 8.8e-17 | 0.9354838709677419 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.79e-48 | 92 | 459 | 58 | 407 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam08305 | NPCBM | 8.82e-39 | 1440 | 1585 | 3 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| PRK10150 | PRK10150 | 3.15e-37 | 93 | 480 | 61 | 442 | beta-D-glucuronidase; Provisional |
| smart00776 | NPCBM | 3.13e-33 | 1436 | 1585 | 1 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| PRK09525 | lacZ | 9.29e-27 | 21 | 459 | 36 | 463 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBA55548.1 | 1.63e-201 | 1670 | 2395 | 23 | 745 |
| BAQ98494.1 | 3.08e-200 | 1670 | 2395 | 23 | 745 |
| AFL04872.1 | 3.08e-200 | 1670 | 2395 | 23 | 745 |
| QRI58269.1 | 3.52e-200 | 1670 | 2395 | 28 | 750 |
| VEG17902.1 | 5.65e-200 | 1670 | 2395 | 46 | 768 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YPJ_A | 5.67e-66 | 43 | 858 | 9 | 807 | ChainA, Beta galactosidase [Niallia circulans],4YPJ_B Chain B, Beta galactosidase [Niallia circulans] |
| 7CWD_A | 3.58e-65 | 43 | 864 | 3 | 807 | ChainA, beta-glalactosidase [Niallia circulans],7CWI_A Chain A, beta-galactosidase [Niallia circulans] |
| 5T98_A | 5.35e-65 | 44 | 851 | 28 | 813 | Crystalstructure of BuGH2Awt [Bacteroides uniformis],5T98_B Crystal structure of BuGH2Awt [Bacteroides uniformis],5T99_A Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis],5T99_B Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 6HPD_A | 1.45e-64 | 34 | 858 | 28 | 804 | Thestructure of a beta-glucuronidase from glycoside hydrolase family 2 [Formosa agariphila KMM 3901] |
| 6B6L_A | 3.52e-59 | 46 | 856 | 10 | 775 | Thecrystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 [Bacteroides cellulosilyticus DSM 14838],6B6L_B The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 [Bacteroides cellulosilyticus DSM 14838] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1V8K7 | 8.58e-154 | 1670 | 2370 | 34 | 726 | Alpha-1,3-galactosidase A OS=Streptacidiphilus griseoplanus OX=66896 GN=glaA PE=1 SV=1 |
| Q826C5 | 3.54e-143 | 1670 | 2287 | 40 | 622 | Alpha-1,3-galactosidase A OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=glaA PE=3 SV=1 |
| P77989 | 2.69e-92 | 41 | 859 | 3 | 739 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| Q8A2Z5 | 6.63e-88 | 1692 | 2223 | 26 | 536 | Alpha-1,3-galactosidase A OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=glaA PE=3 SV=1 |
| Q64MU6 | 1.21e-83 | 1671 | 2223 | 26 | 554 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
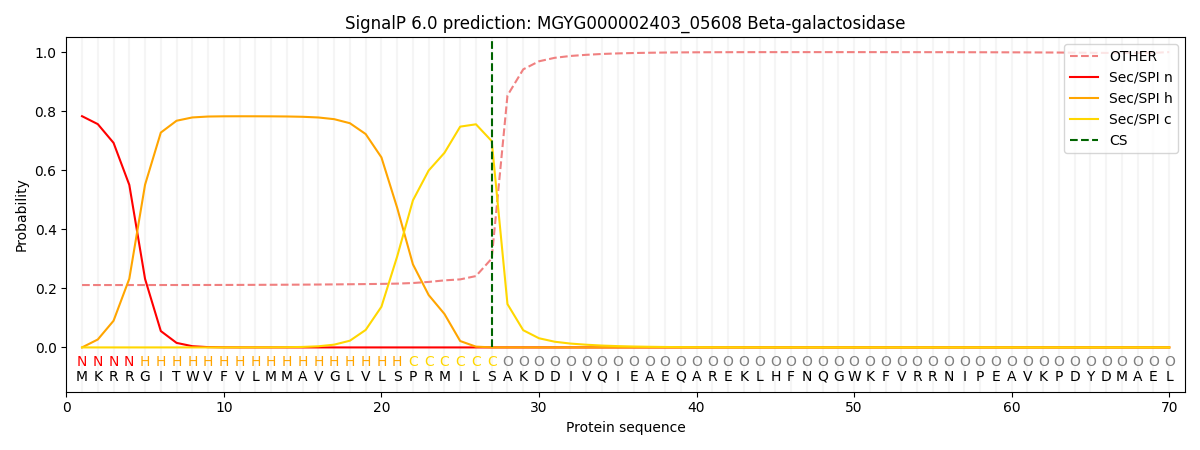
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.229236 | 0.761639 | 0.006942 | 0.001026 | 0.000499 | 0.000637 |