You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002457_02630
You are here: Home > Sequence: MGYG000002457_02630
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
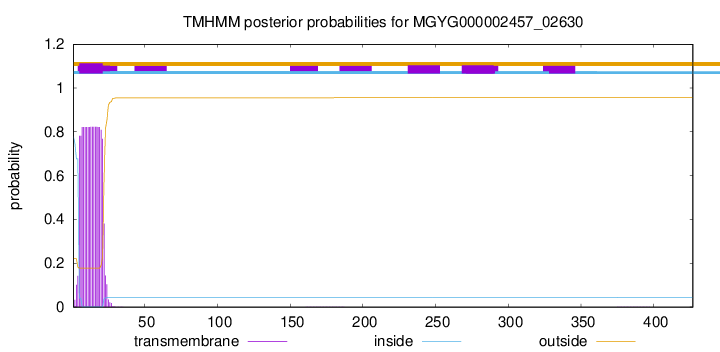
TMHMM annotations
Basic Information help
| Species | Kurthia sp002418445 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_A; Planococcaceae; Kurthia; Kurthia sp002418445 | |||||||||||
| CAZyme ID | MGYG000002457_02630 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 704; End: 1987 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 29 | 384 | 1.1e-128 | 0.9864130434782609 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 1.63e-06 | 139 | 330 | 2 | 148 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 2.09e-06 | 201 | 354 | 2 | 142 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam05048 | NosD | 5.18e-05 | 152 | 386 | 1 | 183 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
| pfam05048 | NosD | 7.16e-05 | 131 | 320 | 26 | 176 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
| COG3420 | NosD | 2.47e-04 | 41 | 391 | 13 | 307 | Nitrous oxidase accessory protein NosD, contains tandem CASH domains [Inorganic ion transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTB20894.1 | 1.24e-145 | 1 | 426 | 1 | 432 |
| QPQ37453.1 | 5.01e-145 | 1 | 424 | 1 | 430 |
| QTB16499.1 | 1.01e-144 | 1 | 426 | 1 | 432 |
| QPA48385.1 | 1.01e-144 | 1 | 426 | 1 | 432 |
| AMO33190.1 | 1.01e-144 | 1 | 426 | 1 | 432 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94576 | 1.65e-101 | 4 | 427 | 7 | 436 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
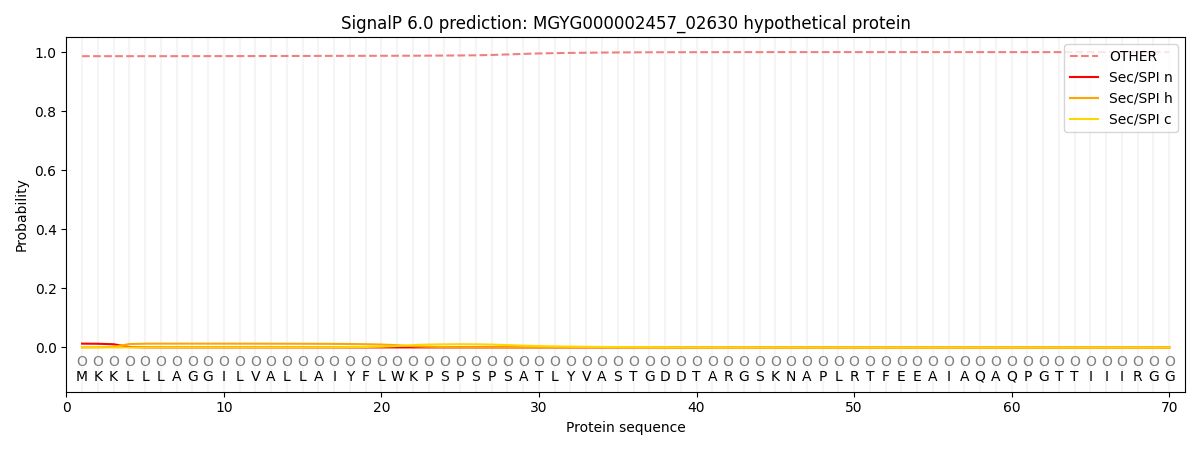
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.986933 | 0.011813 | 0.001146 | 0.000037 | 0.000025 | 0.000072 |