You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002478_00409
You are here: Home > Sequence: MGYG000002478_00409
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola dorei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola dorei | |||||||||||
| CAZyme ID | MGYG000002478_00409 | |||||||||||
| CAZy Family | GH63 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 603433; End: 604815 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH63 | 72 | 445 | 6.9e-20 | 0.3508771929824561 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03200 | Glyco_hydro_63 | 6.65e-12 | 118 | 449 | 126 | 493 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| pfam01204 | Trehalase | 1.54e-11 | 204 | 437 | 284 | 494 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| PRK10137 | PRK10137 | 2.51e-09 | 221 | 405 | 577 | 738 | alpha-glucosidase; Provisional |
| COG3408 | GDB1 | 3.34e-09 | 154 | 420 | 346 | 581 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| pfam06202 | GDE_C | 5.78e-08 | 164 | 379 | 101 | 300 | Amylo-alpha-1,6-glucosidase. This family includes human glycogen branching enzyme AGL. This enzyme contains a number of distinct catalytic activities. It has been shown for the yeast homolog GDB1 that mutations in this region disrupt the enzymes Amylo-alpha-1,6-glucosidase (EC:3.2.1.33). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYN06686.1 | 5.56e-21 | 30 | 441 | 272 | 652 |
| QEW35433.1 | 1.83e-20 | 49 | 441 | 324 | 678 |
| ABR38022.1 | 1.83e-20 | 49 | 441 | 324 | 678 |
| QQY39663.1 | 1.83e-20 | 49 | 441 | 324 | 678 |
| QQY42952.1 | 1.83e-20 | 49 | 441 | 324 | 678 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MHF_A | 3.31e-08 | 349 | 449 | 666 | 776 | Murineendoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_B Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_C Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_D Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus] |
| 4WVA_A | 9.21e-08 | 205 | 441 | 217 | 411 | Crystalstructure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris [Thermus thermophilus HB8],4WVA_B Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris [Thermus thermophilus HB8],4WVB_A Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose [Thermus thermophilus HB8],4WVB_B Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose [Thermus thermophilus HB8],4WVC_A Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate [Thermus thermophilus HB8],4WVC_B Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate [Thermus thermophilus HB8] |
| 2Z07_A | 5.00e-07 | 205 | 441 | 217 | 411 | Crystalstructure of uncharacterized conserved protein from Thermus thermophilus HB8 [Thermus thermophilus],2Z07_B Crystal structure of uncharacterized conserved protein from Thermus thermophilus HB8 [Thermus thermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O88941 | 4.60e-08 | 204 | 449 | 578 | 831 | Mannosyl-oligosaccharide glucosidase OS=Rattus norvegicus OX=10116 GN=Mogs PE=1 SV=1 |
| Q80UM7 | 1.40e-07 | 349 | 449 | 721 | 831 | Mannosyl-oligosaccharide glucosidase OS=Mus musculus OX=10090 GN=Mogs PE=1 SV=1 |
| Q13724 | 3.22e-07 | 349 | 449 | 724 | 834 | Mannosyl-oligosaccharide glucosidase OS=Homo sapiens OX=9606 GN=MOGS PE=1 SV=5 |
| D8QTR2 | 7.10e-06 | 318 | 451 | 367 | 485 | Mannosylglycerate hydrolase MGH1 OS=Selaginella moellendorffii OX=88036 GN=MGH PE=1 SV=1 |
| D8T3S4 | 7.10e-06 | 73 | 451 | 98 | 485 | Mannosylglycerate hydrolase MGH2 OS=Selaginella moellendorffii OX=88036 GN=SELMODRAFT_447962 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
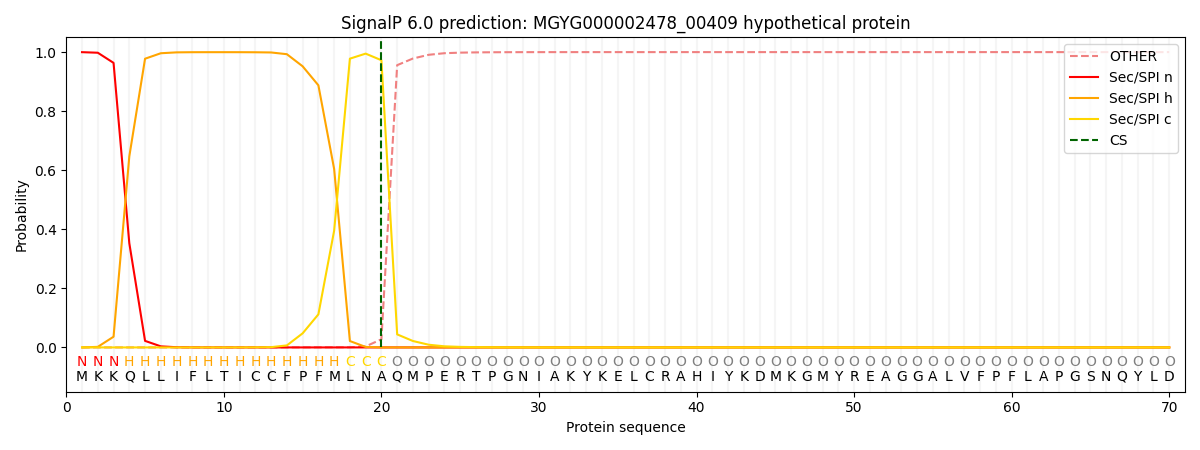
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000296 | 0.998180 | 0.000978 | 0.000184 | 0.000182 | 0.000167 |
