You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002490_00793
You are here: Home > Sequence: MGYG000002490_00793
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
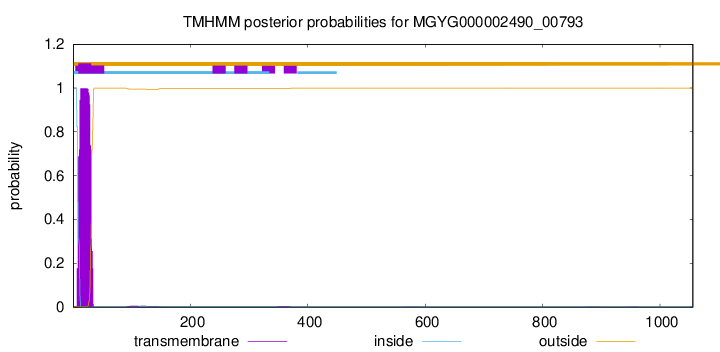
TMHMM annotations
Basic Information help
| Species | Paenibacillus_B thiaminolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_B; Paenibacillus_B thiaminolyticus | |||||||||||
| CAZyme ID | MGYG000002490_00793 | |||||||||||
| CAZy Family | CBM12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 78380; End: 81550 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 190 | 502 | 1.2e-28 | 0.956081081081081 |
| GH18 | 727 | 1037 | 4.4e-25 | 0.9358108108108109 |
| CBM12 | 30 | 63 | 2.5e-16 | 0.9705882352941176 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02871 | GH18_chitinase_D-like | 9.24e-139 | 188 | 506 | 1 | 310 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| cd02871 | GH18_chitinase_D-like | 3.02e-129 | 727 | 1051 | 1 | 312 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| COG3469 | Chi1 | 2.56e-67 | 182 | 512 | 21 | 328 | Chitinase [Carbohydrate transport and metabolism]. |
| COG3469 | Chi1 | 3.14e-49 | 713 | 1037 | 12 | 307 | Chitinase [Carbohydrate transport and metabolism]. |
| pfam00704 | Glyco_hydro_18 | 4.33e-27 | 190 | 494 | 2 | 307 | Glycosyl hydrolases family 18. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDM47453.1 | 0.0 | 487 | 1056 | 1 | 570 |
| ATF10812.1 | 3.03e-217 | 17 | 513 | 17 | 505 |
| VEF91975.1 | 1.26e-216 | 17 | 513 | 17 | 505 |
| QDS36610.1 | 6.84e-216 | 17 | 513 | 17 | 505 |
| AWX53876.1 | 8.69e-214 | 17 | 512 | 17 | 504 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3N11_A | 6.91e-134 | 188 | 513 | 7 | 330 | Crystalstricture of wild-type chitinase from Bacillus cereus NCTU2 [Bacillus cereus],3N12_A Crystal stricture of chitinase in complex with zinc atoms from Bacillus cereus NCTU2 [Bacillus cereus] |
| 3N15_A | 1.92e-133 | 188 | 513 | 7 | 330 | Crystalstricture of E145Q chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
| 5KZ6_A | 4.20e-133 | 188 | 507 | 10 | 327 | 1.25Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis],5KZ6_B 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis] |
| 3N17_A | 7.45e-133 | 188 | 513 | 7 | 330 | Crystalstricture of E145Q/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
| 3N13_A | 1.05e-132 | 188 | 513 | 7 | 330 | Crystalstricture of D143A chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27050 | 2.22e-184 | 28 | 514 | 30 | 516 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| Q05638 | 1.72e-127 | 617 | 1052 | 158 | 596 | Exochitinase 1 OS=Streptomyces olivaceoviridis OX=1921 GN=chi01 PE=1 SV=1 |
| D4AVJ0 | 3.76e-102 | 745 | 1053 | 17 | 329 | Probable class II chitinase ARB_00204 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_00204 PE=1 SV=2 |
| A5FB63 | 4.89e-61 | 94 | 497 | 1046 | 1465 | Chitinase ChiA OS=Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / NBRC 14942 / NCIMB 11054 / UW101) OX=376686 GN=chiA PE=1 SV=1 |
| Q838S2 | 8.35e-34 | 727 | 1037 | 42 | 325 | Chitinase OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=EF_0361 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
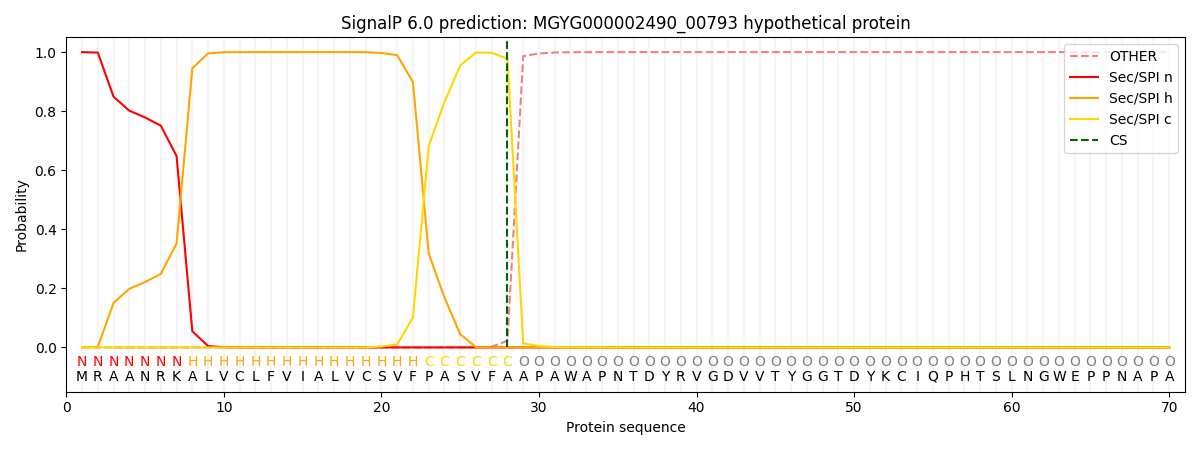
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000208 | 0.999153 | 0.000168 | 0.000177 | 0.000149 | 0.000132 |